SARS-CoV-2 Rapid Antigen Test Based on a New Anti-Nucleocapsid Protein Monoclonal Antibody: Development and Real-Time Validation
- PMID: 37894080
- PMCID: PMC10608853
- DOI: 10.3390/microorganisms11102422
SARS-CoV-2 Rapid Antigen Test Based on a New Anti-Nucleocapsid Protein Monoclonal Antibody: Development and Real-Time Validation
Abstract
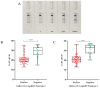
SARS-CoV-2 diagnostic tests have become an important tool for pandemic control. Among the alternatives for COVID-19 diagnosis, antigen rapid diagnostic tests (Ag-RDT) are very convenient and widely used. However, as SARS-CoV-2 variants may continuously emerge, the replacement of tests and reagents may be required to maintain the sensitivity of Ag-RDTs. Here, we describe the development and validation of an Ag-RDT during an outbreak of the Omicron variant, including the characterization of a new monoclonal antibody (anti-DTC-N 1B3 mAb) that recognizes the Nucleocapsid protein (N). The anti-DTC-N 1B3 mAb recognized the sequence TFPPTEPKKDKKK located at the C-terminus of the N protein of main SARS-CoV-2 variants of concern. Accordingly, the Ag-RDT prototypes using the anti-DTC-N 1B3 mAB detected all the SARS-CoV-2 variants-Wuhan, Alpha, Gamma, Delta, P2 and Omicron. The performance of the best prototype (sensitivity of 95.2% for samples with Ct ≤ 25; specificity of 98.3% and overall accuracy of 85.0%) met the WHO recommendations. Moreover, results from a patients' follow-up study indicated that, if performed within the first three days after onset of symptoms, the Ag-RDT displayed 100% sensitivity. Thus, the new mAb and the Ag-RDT developed herein may constitute alternative tools for COVID-19 point-of-care diagnosis and epidemiological surveillance.
Keywords: Ag-RDT development; IgG2b monoclonal antibody; SARS-CoV-2; diagnosis; follow-up study; nucleocapsid (N) antigen; validation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- WHO Coronavirus (COVID-19) Dashboard. [(accessed on 4 September 2023)]. Available online: https://covid19.who.int/
-
- Giovanetti M., Fonseca V., Wilkinson E., Tegally H., San E.J., Althaus C.L., Xavier J., Nanev Slavov S., Viala V.L., Ranieri Jerônimo Lima A. Replacement of the Gamma by the Delta Variant in Brazil: Impact of Lineage Displacement on the Ongoing Pandemic. Virus Evol. 2022;8:veac024. doi: 10.1093/ve/veac024. - DOI - PMC - PubMed
-
- Naveca F.G., Nascimento V., Souza V., Corado A.L., Nascimento F., Silva G., Mejía M.C., Brandão M.J., Costa Á., Duarte D. Spread of Gamma (P. 1) Sub-Lineages Carrying Spike Mutations Close to the Furin Cleavage Site and Deletions in the N-Terminal Domain Drives Ongoing Transmission of SARS-CoV-2 in Amazonas, Brazil. medRxiv. 2021;10:e02366-21. doi: 10.1128/spectrum.02366-21. - DOI - PMC - PubMed
-
- Baral P., Bhattarai N., Hossen M.L., Stebliankin V., Gerstman B.S., Narasimhan G., Chapagain P.P. Mutation-Induced Changes in the Receptor-Binding Interface of the SARS-CoV-2 Delta Variant B. 1.617. 2 and Implications for Immune Evasion. Biochem. Biophys. Res. Commun. 2021;574:14–19. doi: 10.1016/j.bbrc.2021.08.036. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

