Unveiling Rare Pathogens and Antibiotic Resistance in Tanzanian Cholera Outbreak Waters
- PMID: 37894148
- PMCID: PMC10609457
- DOI: 10.3390/microorganisms11102490
Unveiling Rare Pathogens and Antibiotic Resistance in Tanzanian Cholera Outbreak Waters
Abstract
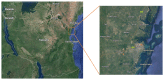
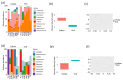
The emergence of antibiotic resistance is a global health concern. Therefore, understanding the mechanisms of its spread is crucial for implementing evidence-based strategies to tackle resistance in the context of the One Health approach. In developing countries where sanitation systems and access to clean and safe water are still major challenges, contamination may introduce bacteria and bacteriophages harboring antibiotic resistance genes (ARGs) into the environment. This contamination can increase the risk of exposure and community transmission of ARGs and infectious pathogens. However, there is a paucity of information on the mechanisms of bacteriophage-mediated spread of ARGs and patterns through the environment. Here, we deploy Droplet Digital PCR (ddPCR) and metagenomics approaches to analyze the abundance of ARGs and bacterial pathogens disseminated through clean and wastewater systems. We detected a relatively less-studied and rare human zoonotic pathogen, Vibrio metschnikovii, known to spread through fecal--oral contamination, similarly to V. cholerae. Several antibiotic resistance genes were identified in both bacterial and bacteriophage fractions from water sources. Using metagenomics, we detected several resistance genes related to tetracyclines and beta-lactams in all the samples. Environmental samples from outlet wastewater had a high diversity of ARGs and contained high levels of blaOXA-48. Other identified resistance profiles included tetA, tetM, and blaCTX-M9. Specifically, we demonstrated that blaCTX-M1 is enriched in the bacteriophage fraction from wastewater. In general, however, the bacterial community has a significantly higher abundance of resistance genes compared to the bacteriophage population. In conclusion, the study highlights the need to implement environmental monitoring of clean and wastewater to inform the risk of infectious disease outbreaks and the spread of antibiotic resistance in the context of One Health.
Keywords: ESBL; One Health; antibiotic resistance; bacteriophage; cholera; microbiota; transduction; water; zoonotic pathogen.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Ministry of Agriculture, Livestock and Fisheries . National Action Plan for Antimicrobial Resistance 2017–2022. Volume 136 Ministry of Agriculture; Dar es Salaam, Tanzania: 2017.
-
- Ministry of Health . United Republic of Tanzania: The National Action Plan on Antimicrobial Resistance 2023–2028. Ministry of Health; Dodoma, Tanzania: 2022. p. 136.
Grants and funding
LinkOut - more resources
Full Text Sources

