The Carcinogenic Potential of Bisphenol A in the Liver Based on Transcriptomic Studies
- PMID: 37894381
- PMCID: PMC10605469
- DOI: 10.3390/cancers15205014
The Carcinogenic Potential of Bisphenol A in the Liver Based on Transcriptomic Studies
Abstract
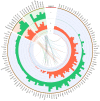
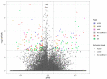
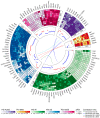
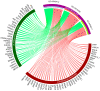
Bisphenol A (BPA) is an environmental toxin widely used in the production of polycarbonate plastics. A correlation exists between BPA tissue contamination and the occurrence of pathological conditions, including cancer. First-passage detoxification of high BPA amounts in the liver promotes hepatotoxicity and morphological alterations of this organ, but there is a lack of knowledge about the molecular mechanisms underlying these phenomena. This prompted us to investigate changes in the liver transcriptomics of 3-month-old female mice exposed to BPA (50 mg/kg) in drinking water for 3 months. Five female mice served as controls. The animals were euthanized, the livers were collected, and RNA was extracted to perform RNA-seq analysis. The multistep transcriptomic bioinformatics revealed 120 differentially expressed genes (DEGs) in the BPA-exposed samples. Gene Ontology (GO) annotations indicated that DEGs have been assigned to many biological processes, including "macromolecule modification" and "protein metabolic process". Several of the revealed DEGs have been linked to the pathogenesis of severe metabolic liver disorders and malignant tumors, in particular hepatocellular carcinoma. Data from this study suggest that BPA has a significant impact on gene expression in the liver, which is predictive of the carcinogenic potential of this compound in this organ.
Keywords: RNA-Seq; bisphenol A; hepatocellular carcinoma; liver; mitochondrial dysfunction.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Grants and funding
LinkOut - more resources
Full Text Sources

