Mucosal Microbiome in Patients with Early Bowel Polyps: Inferences from Short-Read and Long-Read 16S rRNA Sequencing
- PMID: 37894412
- PMCID: PMC10605900
- DOI: 10.3390/cancers15205045
Mucosal Microbiome in Patients with Early Bowel Polyps: Inferences from Short-Read and Long-Read 16S rRNA Sequencing
Abstract
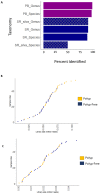
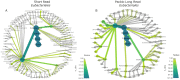
Numerous studies have correlated dysbiosis in stool microbiota with colorectal cancer (CRC); however, fewer studies have investigated the mucosal microbiome in pre-cancerous bowel polyps. The short-read sequencing of variable regions in the 16S rRNA gene has commonly been used to infer bacterial taxonomy, and this has led, in part, to inconsistent findings between studies. Here, we examined mucosal microbiota from patients who presented with one or more polyps, compared to patients with no polyps, at the time of colonoscopy. We evaluated the results obtained using both short-read and PacBio long-read 16S rRNA sequencing. Neither sequencing technology identified significant differences in microbial diversity measures between patients with or without bowel polyps. Differential abundance measures showed that amplicon sequence variants (ASVs) associated with Ruminococcus gnavus and Escherichia coli were elevated in mucosa from polyp patients, while ASVs associated with Parabacteroides merdae, Veillonella nakazawae, and Sutterella wadsworthensis were relatively decreased. Only R. gnavus was consistently identified using both sequencing technologies as being altered between patients with polyps compared to patients without polyps, suggesting differences in technologies and bioinformatics processing impact study findings. Several of the differentially abundant bacteria identified using either sequencing technology are associated with inflammatory bowel diseases despite these patients being excluded from the current study, which suggests that early bowel neoplasia may be associated with a local inflammatory niche.
Keywords: 16S rRNA sequencing; PacBio long-read sequencing; bowel polyps; gut microbiome.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures






References
-
- Abed J., Emgard J.E., Zamir G., Faroja M., Almogy G., Grenov A., Sol A., Naor R., Pikarsky E., Atlan K.A., et al. Fap2 Mediates Fusobacterium nucleatum Colorectal Adenocarcinoma Enrichment by Binding to Tumor-Expressed Gal-GalNAc. Cell Host Microbe. 2016;20:215–225. doi: 10.1016/j.chom.2016.07.006. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

