Revealing Allosteric Mechanism of Amino Acid Binding Proteins from Open to Closed State
- PMID: 37894619
- PMCID: PMC10609312
- DOI: 10.3390/molecules28207139
Revealing Allosteric Mechanism of Amino Acid Binding Proteins from Open to Closed State
Abstract
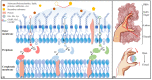
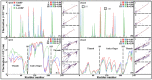
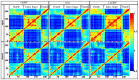
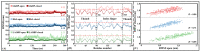
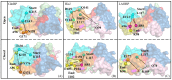
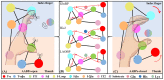
Amino acid binding proteins (AABPs) undergo significant conformational closure in the periplasmic space of Gram-negative bacteria, tightly binding specific amino acid substrates and then initiating transmembrane transport of nutrients. Nevertheless, the possible closure mechanisms after substrate binding, especially long-range signaling, remain unknown. Taking three typical AABPs-glutamine binding protein (GlnBP), histidine binding protein (HisJ) and lysine/arginine/ornithine binding protein (LAOBP) in Escherichia coli (E. coli)-as research subjects, a series of theoretical studies including sequence alignment, Gaussian network model (GNM), anisotropic network model (ANM), conventional molecular dynamics (cMD) and neural relational inference molecular dynamics (NRI-MD) simulations were carried out. Sequence alignment showed that GlnBP, HisJ and LAOBP have high structural similarity. According to the results of the GNM and ANM, AABPs' Index Finger and Thumb domains exhibit closed motion tendencies that contribute to substrate capture and stable binding. Based on cMD trajectories, the Index Finger domain, especially the I-Loop region, exhibits high molecular flexibility, with residues 11 and 117 both being potentially key residues for receptor-ligand recognition and initiation of receptor allostery. Finally, the signaling pathway of AABPs' conformational closure was revealed by NRI-MD training and trajectory reconstruction. This work not only provides a complete picture of AABPs' recognition mechanism and possible conformational closure, but also aids subsequent structure-based design of small-molecule oncology drugs.
Keywords: Gaussian network model; allosteric mechanism; amino acid binding protein; anisotropic network model; neural relational inference molecular dynamics.
Conflict of interest statement
The authors declare no conflict of interest, financial or otherwise.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

