Comparison of Methods for Quantifying Extracellular Vesicles of Gram-Negative Bacteria
- PMID: 37894776
- PMCID: PMC10606555
- DOI: 10.3390/ijms242015096
Comparison of Methods for Quantifying Extracellular Vesicles of Gram-Negative Bacteria
Abstract
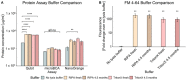
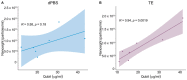
There are a variety of methods employed by laboratories for quantifying extracellular vesicles isolated from bacteria. As a result, the ability to compare results across published studies can lead to questions regarding the suitability of methods and buffers for accurately quantifying these vesicles. Within the literature, there are several common methods for vesicle quantification. These include lipid quantification using the lipophilic dye FM 4-64, protein quantification using microBCA, Qubit, and NanoOrange assays, or direct vesicle enumeration using nanoparticle tracking analysis. In addition, various diluents and lysis buffers are also used to resuspend and treat vesicles. In this study, we directly compared the quantification of a bacterial outer membrane vesicle using several commonly used methods. We also tested the impact of different buffers, buffer age, lysis method, and vesicle diluent on vesicle quantification. The results showed that buffer age had no significant effect on vesicle quantification, but the lysis method impacted the reliability of measurements using Qubit and NanoOrange. The microBCA assay displayed the least variability in protein concentration values and was the most consistent, regardless of the buffer or diluent used. MicroBCA also demonstrated the strongest correlation to the NTA-determined particle number across a range of vesicle concentrations. Overall, these results indicate that with appropriate diluent and buffer choice, microBCA vs. NTA standard curves could be generated and the microBCA assay used to estimate the particle number when NTA instrumentation is not readily available.
Keywords: FM 4-64; NTA; NanoOrange; Qubit; bacterial extracellular vesicle; microBCA; nanoparticle tracking analysis; outer membrane vesicles; vesicle quantification.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

