AZD-7648, a DNA-PK Inhibitor, Induces DNA Damage, Apoptosis, and Cell Cycle Arrest in Chronic and Acute Myeloid Leukemia Cells
- PMID: 37895013
- PMCID: PMC10607085
- DOI: 10.3390/ijms242015331
AZD-7648, a DNA-PK Inhibitor, Induces DNA Damage, Apoptosis, and Cell Cycle Arrest in Chronic and Acute Myeloid Leukemia Cells
Abstract
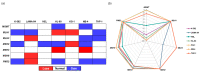
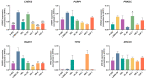
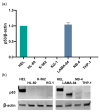
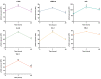
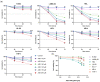
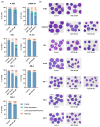
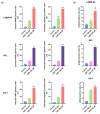
The non-homologous end joining pathway is vital for repairing DNA double-strand breaks (DSB), with DNA-dependent protein kinase (DNA-PK) playing a critical role. Altered DNA damage response (DDR) in chronic (CML) and acute myeloid leukemia (AML) offers potential therapeutic opportunities. We studied the therapeutic potential of AZD-7648 (DNA-PK inhibitor) in CML and AML cell lines. This study used two CML (K-562 and LAMA-84) and five AML (HEL, HL-60, KG-1, NB-4, and THP-1) cell lines. DDR gene mutations were obtained from the COSMIC database. The copy number and methylation profile were evaluated using MS-MLPA and DDR genes, and telomere length using qPCR. p53 protein expression was assessed using Western Blot, chromosomal damage through cytokinesis-block micronucleus assay, and γH2AX levels and DSB repair kinetics using flow cytometry. Cell density and viability were analyzed using trypan blue assay after treatment with AZD-7648 in concentrations ranging from 10 to 200 µM. Cell death, cell cycle distribution, and cell proliferation rate were assessed using flow cytometry. The cells displayed different DNA baseline damage, DDR gene expressions, mutations, genetic/epigenetic changes, and p53 expression. Only HEL cells displayed inefficient DSB repair. The LAMA-84, HEL, and KG-1 cells were the most sensitive to AZD-7648, whereas HL-60 and K-562 showed a lower effect on density and viability. Besides the reduction in cell proliferation, AZD-7648 induced apoptosis, cell cycle arrest, and DNA damage. In conclusion, these results suggest that AZD-7648 holds promise as a potential therapy for myeloid leukemias, however, with variations in drug sensitivity among tested cell lines, thus supporting further investigation to identify the specific factors influencing sensitivity to this DNA-PK inhibitor.
Keywords: AZD-7648; DNA damage repair; DNA-PK inhibitor; myeloid leukemia; therapeutic target.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures
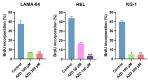
Similar articles
-
Wee1 promotes cell proliferation and imatinib resistance in chronic myeloid leukemia via regulating DNA damage repair dependent on ATM-γH2AX-MDC1.Cell Commun Signal. 2022 Dec 27;20(1):199. doi: 10.1186/s12964-022-01021-z. Cell Commun Signal. 2022. PMID: 36575478 Free PMC article.
-
DNA-PK inhibitor peposertib enhances p53-dependent cytotoxicity of DNA double-strand break inducing therapy in acute leukemia.Sci Rep. 2021 Jun 9;11(1):12148. doi: 10.1038/s41598-021-90500-3. Sci Rep. 2021. PMID: 34108527 Free PMC article.
-
DNA-dependent protein kinase: effect on DSB repair, G2/M checkpoint and mode of cell death in NSCLC cell lines.Int J Radiat Biol. 2019 Sep;95(9):1205-1219. doi: 10.1080/09553002.2019.1642536. Epub 2019 Jul 24. Int J Radiat Biol. 2019. PMID: 31287365
-
[Analysis of the role of DNA repair, regulation of cell cycle and apoptosis in the radiation-induced adaptive response of mammalian cells].Radiats Biol Radioecol. 2003 Jan-Feb;43(1):19-28. Radiats Biol Radioecol. 2003. PMID: 12677654 Review. Russian.
-
Involvement of DNA-PK and ATM in radiation- and heat-induced DNA damage recognition and apoptotic cell death.J Radiat Res. 2010;51(5):493-501. doi: 10.1269/jrr.10039. Epub 2010 Aug 28. J Radiat Res. 2010. PMID: 20814172 Review.
Cited by
-
DNA Damage, DNA Repair, and Cancer: Second Edition.Int J Mol Sci. 2023 Nov 28;24(23):16835. doi: 10.3390/ijms242316835. Int J Mol Sci. 2023. PMID: 38069158 Free PMC article.
-
N6-methyladenosine binding protein YTHDF2 inhibits gastric cancer cell growth and predicts better prognosis in patients with gastric cancer.Transl Oncol. 2025 Jun;56:102395. doi: 10.1016/j.tranon.2025.102395. Epub 2025 Apr 11. Transl Oncol. 2025. PMID: 40215678 Free PMC article.
-
Medicinal chemistry breakthroughs on ATM, ATR, and DNA-PK inhibitors as prospective cancer therapeutics.J Enzyme Inhib Med Chem. 2025 Dec;40(1):2489720. doi: 10.1080/14756366.2025.2489720. Epub 2025 Apr 21. J Enzyme Inhib Med Chem. 2025. PMID: 40256842 Free PMC article. Review.
References
-
- Lam F.C. The DNA damage response—From cell biology to human disease. J. Transl. Genet. Genom. 2022;6:204–222. doi: 10.20517/jtgg.2021.61. - DOI
-
- Fok J.H.L., Ramos-Montoya A., Vazquez-Chantada M., Wijnhoven P.W.G., Follia V., James N., Farrington P.M., Karmokar A., Willis S.E., Cairns J., et al. AZD7648 is a potent and selective DNA-PK inhibitor that enhances radiation, chemotherapy and olaparib activity. Nat. Commun. 2019;10:5065. doi: 10.1038/s41467-019-12836-9. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- POCI-01-0145-FEDER-007440/Fundação para a Ciência e Tecnologia
- NA/Liga Portuguesa Contra o Cancro - Núcleo Regional do Centro
- NA/Sociedade Portuguesa de Hematologia
- UID/NEU/04539/2019, UIDB/04539/2020, UIDP/04539/2020/Fundação para a Ciência e Tecnologia
- 2020.08261.BD, 2022.10127.BD, 2020.07672.BD and SFRH/BD/145531/2019/Fundação para a Ciência e Tecnologia
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

