Tissue-Specific Tumour Suppressor and Oncogenic Activities of the Polycomb-like Protein MTF2
- PMID: 37895228
- PMCID: PMC10606531
- DOI: 10.3390/genes14101879
Tissue-Specific Tumour Suppressor and Oncogenic Activities of the Polycomb-like Protein MTF2
Abstract
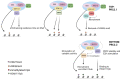
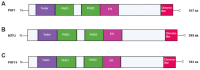
The Polycomb repressive complex 2 (PRC2) is a conserved chromatin-remodelling complex that catalyses the trimethylation of histone H3 lysine 27 (H3K27me3), a mark associated with gene silencing. PRC2 regulates chromatin structure and gene expression during organismal and tissue development and tissue homeostasis in the adult. PRC2 core subunits are associated with various accessory proteins that modulate its function and recruitment to target genes. The multimeric composition of accessory proteins results in two distinct variant complexes of PRC2, PRC2.1 and PRC2.2. Metal response element-binding transcription factor 2 (MTF2) is one of the Polycomb-like proteins (PCLs) that forms the PRC2.1 complex. MTF2 is highly conserved, and as an accessory subunit of PRC2, it has important roles in embryonic stem cell self-renewal and differentiation, development, and cancer progression. Here, we review the impact of MTF2 in PRC2 complex assembly, catalytic activity, and spatiotemporal function. The emerging paradoxical evidence suggesting that MTF2 has divergent roles as either a tumour suppressor or an oncogene in different tissues merits further investigations. Altogether, our review illuminates the context-dependent roles of MTF2 in Polycomb group (PcG) protein-mediated epigenetic regulation. Its impact on disease paves the way for a deeper understanding of epigenetic regulation and novel therapeutic strategies.
Keywords: MTF2; PHF1; PHF19; Polycomb repressive complex 2; Polycomb-like; cancer; development; epigenetics; oncogene; tumour suppressor.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Polycomb-like Proteins in Gene Regulation and Cancer.Genes (Basel). 2023 Apr 18;14(4):938. doi: 10.3390/genes14040938. Genes (Basel). 2023. PMID: 37107696 Free PMC article. Review.
-
Distinct PRC2 subunits regulate maintenance and establishment of Polycomb repression during differentiation.Mol Cell. 2021 Jun 17;81(12):2625-2639.e5. doi: 10.1016/j.molcel.2021.03.038. Epub 2021 Apr 21. Mol Cell. 2021. PMID: 33887196 Free PMC article.
-
MTF2 recruits Polycomb Repressive Complex 2 by helical-shape-selective DNA binding.Nat Genet. 2018 Jul;50(7):1002-1010. doi: 10.1038/s41588-018-0134-8. Epub 2018 May 28. Nat Genet. 2018. PMID: 29808031
-
Kicking against the PRCs - A Domesticated Transposase Antagonises Silencing Mediated by Polycomb Group Proteins and Is an Accessory Component of Polycomb Repressive Complex 2.PLoS Genet. 2015 Dec 7;11(12):e1005660. doi: 10.1371/journal.pgen.1005660. eCollection 2015 Dec. PLoS Genet. 2015. PMID: 26642436 Free PMC article.
-
Polycomb repressive complex 2 in embryonic stem cells: an overview.Protein Cell. 2010 Dec;1(12):1056-62. doi: 10.1007/s13238-010-0142-7. Epub 2011 Jan 8. Protein Cell. 2010. PMID: 21213100 Free PMC article. Review.
Cited by
-
Addressing the mean-variance relationship in spatially resolved transcriptomics data with spoon.bioRxiv [Preprint]. 2024 Nov 8:2024.11.04.621867. doi: 10.1101/2024.11.04.621867. bioRxiv. 2024. Update in: Biostatistics. 2024 Dec 31;26(1):kxaf012. doi: 10.1093/biostatistics/kxaf012. PMID: 39574747 Free PMC article. Updated. Preprint.
-
MTF2 facilitates the advancement of osteosarcoma through mediating EZH2/SFRP1/Wnt signaling.J Orthop Surg Res. 2024 Aug 8;19(1):467. doi: 10.1186/s13018-024-04965-9. J Orthop Surg Res. 2024. PMID: 39118123 Free PMC article.
-
The PRC2.1 Subcomplex Opposes G1 Progression through Regulation of CCND1 and CCND2.bioRxiv [Preprint]. 2024 Oct 16:2024.03.18.585604. doi: 10.1101/2024.03.18.585604. bioRxiv. 2024. Update in: Elife. 2025 Feb 04;13:RP97577. doi: 10.7554/eLife.97577. PMID: 38562687 Free PMC article. Updated. Preprint.
-
The PRC2.1 subcomplex opposes G1 progression through regulation of CCND1 and CCND2.Elife. 2025 Feb 4;13:RP97577. doi: 10.7554/eLife.97577. Elife. 2025. PMID: 39903505 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

