Evaluation of Alectinib Metabolic Stability in HLMs Using Fast LC-MS/MS Method: In Silico ADME Profile, P450 Metabolic Lability, and Toxic Alerts Screening
- PMID: 37896209
- PMCID: PMC10610548
- DOI: 10.3390/pharmaceutics15102449
Evaluation of Alectinib Metabolic Stability in HLMs Using Fast LC-MS/MS Method: In Silico ADME Profile, P450 Metabolic Lability, and Toxic Alerts Screening
Abstract
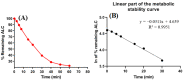
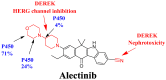
Alectinib, also known as Alecensa®, is prescribed for the therapeutic treatment of individuals diagnosed with metastatic non-small cell lung cancer (NSCLC) who have a specific genetic mutation referred to as anaplastic lymphoma kinase (ALK) positivity. The Food and Drug Administration granted regular approval to alectinib, a drug developed by Hoffmann-La Roche, Inc. (Basel, Switzerland)/Genentech, Inc. (South San Francisco, CA, USA), on 6 November 2017. The screening of the metabolic stability and identification of hazardous alarms within the chemical structure of ALC was conducted using the StarDrop software package (version 6.6), which incorporates the P450 metabolic module and DEREK software (KB 2018 1.1). The primary aim of this investigation was to develop a high-throughput and accurate LC-MS/MS technique for the quantification of ALC in the metabolic matrix (human liver microsomes; HLMs). The aforementioned methodology was subsequently employed to assess the metabolic stability of ALC in HLMs through in vitro tests, with the obtained results further validated using in silico software. The calibration curve of the ALC showed a linear correlation that exists within the concentration range from 1 to 3000 ng/mL. The LC-MS/MS approach that was recommended exhibited accuracy and precision levels for both inter-day and intra-day measurements. Specifically, the accuracy values ranged from -2.56% to 3.45%, while the precision values ranged from -3.78% to 4.33%. The sensitivity of the established approach was proved by its ability to adhere to an LLOQ of 0.82 ng/mL. The half-life (t1/2) and intrinsic clearance (Clint) of ALC were estimated to be 22.28 min and 36.37 mL/min/kg, correspondingly, using in vitro experiments. The ALC exhibited a moderate extraction ratio. The metabolic stability and safety properties of newly created derivatives can be enhanced by making modest adjustments to the morpholine and piperidine rings or by substituting the substituent, as per computational software. In in silico ADME prediction, ALC was shown to have poor water solubility and high gastrointestinal absorption along with inhibition of some cytochrome P450s (CYP2C19 and CYP2C9) without inhibition of others (CYP1A2, CYP3A4, and CYP2D6) and P-glycoprotein substrate. The study design that involves using both laboratory experiments and different in silico software demonstrates a novel and groundbreaking approach in the establishment and uniformization of LC-MS/MS techniques for the estimation of ALC concentrations, identifying structural alerts and the assessment of its metabolic stability. The utilization of this study strategy has the potential to be employed in the screening and optimization of prospective compounds during the drug creation process. This strategy may also facilitate the development of novel derivatives of the medicine that maintain the same biological action by targeted structural modifications, based on an understanding of the structural alerts included within the chemical structure of ALC.
Keywords: ADME profile; DEREK software; LC-MS/MS approach; P450 metabolic mode; StarDrop software; alectinib; greenness; in vitro half-life; intrinsic clearance; metabolic stability.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Assessment of the in vitro metabolic stability of CEP-37440, a selective FAK/ALK inhibitor, in HLMs using fast UPLC-MS/MS method: in silico metabolic lability and DEREK alerts screening.Front Chem. 2024 Sep 26;12:1323738. doi: 10.3389/fchem.2024.1323738. eCollection 2024. Front Chem. 2024. PMID: 39391832 Free PMC article.
-
Characterization of the in vitro metabolic profile of nazartinib in HLMs using UPLC-MS/MS method: In silico metabolic lability and DEREK structural alerts screening using StarDrop software.Heliyon. 2024 Jul 5;10(13):e34109. doi: 10.1016/j.heliyon.2024.e34109. eCollection 2024 Jul 15. Heliyon. 2024. PMID: 39091946 Free PMC article.
-
An Ultrafast UPLC-MS/MS Method for Characterizing the In Vitro Metabolic Stability of Acalabrutinib.Molecules. 2023 Oct 23;28(20):7220. doi: 10.3390/molecules28207220. Molecules. 2023. PMID: 37894699 Free PMC article.
-
Safety and nutritional assessment of GM plants and derived food and feed: the role of animal feeding trials.Food Chem Toxicol. 2008 Mar;46 Suppl 1:S2-70. doi: 10.1016/j.fct.2008.02.008. Epub 2008 Feb 13. Food Chem Toxicol. 2008. PMID: 18328408 Review.
-
Methodologies for investigating drug metabolism at the early drug discovery stage: prediction of hepatic drug clearance and P450 contribution.Curr Drug Metab. 2010 Oct;11(8):678-85. doi: 10.2174/138920010794233503. Curr Drug Metab. 2010. PMID: 20973757 Review.
Cited by
-
Synthesis of Co/Co3O4 Heterostructure in N-Doped Porous, Amorphous Carbon: A Superior Electrochemical Sensor for Sensitive Determination of Alectinib in Various Fluids.ACS Omega. 2024 Oct 24;9(44):44282-44292. doi: 10.1021/acsomega.4c04821. eCollection 2024 Nov 5. ACS Omega. 2024. PMID: 39524646 Free PMC article.
-
3D spheroid HepaRG and fluorescent biphasic tracer for CYP3A4-mediated antibiotic interaction monitoring in sepsis.Anal Bioanal Chem. 2024 Aug;416(19):4261-4274. doi: 10.1007/s00216-024-05363-0. Epub 2024 Jun 6. Anal Bioanal Chem. 2024. PMID: 38839687
References
Grants and funding
LinkOut - more resources
Full Text Sources

