A Single-Dose Intramuscular Immunization of Pigs with Lipid Nanoparticle DNA Vaccines Based on the Hemagglutinin Antigen Confers Complete Protection against Challenge Infection with the Homologous Influenza Virus Strain
- PMID: 37896997
- PMCID: PMC10611089
- DOI: 10.3390/vaccines11101596
A Single-Dose Intramuscular Immunization of Pigs with Lipid Nanoparticle DNA Vaccines Based on the Hemagglutinin Antigen Confers Complete Protection against Challenge Infection with the Homologous Influenza Virus Strain
Abstract
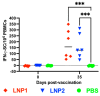
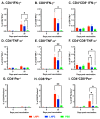
The Influenza A virus of swine (IAV-S) is highly prevalent and causes significant economic losses to swine producers. Due to the highly variable and rapidly evolving nature of the virus, it is critical to develop a safe and versatile vaccine platform that allows for frequent updates of the vaccine immunogens to cope with the emergence of new viral strains. The main objective of this study was to assess the feasibility of using lipid nanoparticles (LNPs) as nanocarriers for delivering DNA plasmid encoding the viral hemagglutinin (HA) gene in pigs. The intramuscular administration of a single dose of the LNP-DNA vaccines resulted in robust systemic and mucosal responses in pigs. Importantly, the vaccinated pigs were fully protected against challenge infection with the homologous IAV-S strain, with only 1 out of 12 vaccinated pigs shedding a low amount of viral genomic RNA in its nasal cavity. No gross or microscopic lesions were observed in the lungs of the vaccinated pigs at necropsy. Thus, the LNP-DNA vaccines are highly effective in protecting pigs against the homologous IAV-S strain and can serve as a promising platform for the rapid development of IAV-S vaccines.
Keywords: DNA vaccine; lipid nanoparticles; swine influenza virus; vaccine platform.
Conflict of interest statement
H.L.X.V., T.N.N., and S.S. have filed a patent application entitled “Methods and Compositions for Vaccine Development and Delivery”. The other authors declare no conflicts of interest.
Figures




References
-
- Anderson T.K., Macken C.A., Lewis N.S., Scheuermann R.H., Van Reeth K., Brown I.H., Swenson S.L., Simon G., Saito T., Berhane Y., et al. A Phylogeny-Based Global Nomenclature System and Automated Annotation Tool for H1 Hemagglutinin Genes from Swine Influenza A Viruses. mSphere. 2016;1:e00275-16. doi: 10.1128/mSphere.00275-16. - DOI - PMC - PubMed
-
- Venkatesh D., Anderson T.K., Kimble J.B., Chang J., Lopes S., Souza C.K., Pekosz A., Shaw-Saliba K., Rothman R.E., Chen K.F., et al. Antigenic Characterization and Pandemic Risk Assessment of North American H1 Influenza A Viruses Circulating in Swine. Microbiol. Spectr. 2022;10:e0178122. doi: 10.1128/spectrum.01781-22. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

