Enhancing wheat regeneration and genetic transformation through overexpression of TaLAX1
- PMID: 37897039
- PMCID: PMC11121199
- DOI: 10.1016/j.xplc.2023.100738
Enhancing wheat regeneration and genetic transformation through overexpression of TaLAX1
Abstract
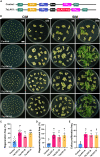
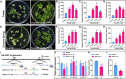
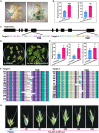
In the realm of genetically transformed crops, the process of plant regeneration holds utmost significance. However, the low regeneration efficiency of several wheat varieties currently restricts the use of genetic transformation for gene functional analysis and improved crop production. This research explores overexpression of TaLAX PANICLE1 (TaLAX1), which markedly enhances regeneration efficiency, thereby boosting genetic transformation and genome editing in wheat. Particularly noteworthy is the substantial increase in regeneration efficiency of common wheat varieties previously regarded as recalcitrant to genetic transformation. Our study shows that increased expression of TaGROWTH-REGULATING FACTOR (TaGRF) genes, alongside that of their co-factor, TaGRF-INTERACTING FACTOR 1 (TaGIF1), enhances cytokinin accumulation and auxin response, which may play pivotal roles in the improved regeneration and transformation of TaLAX1-overexpressing wheat plants. Overexpression of TaLAX1 homologs also significantly increases the regeneration efficiency of maize and soybean, suggesting that both monocot and dicot crops can benefit from this enhancement. Our findings shed light on a gene that enhances wheat genetic transformation and elucidate molecular mechanisms that potentially underlie wheat regeneration.
Keywords: TaGRF4–TaGIF1; TaLAX1; auxin; cytokinin; genetic transformation; plant regeneration; wheat.
Copyright © 2023 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures




References
-
- Bouchabke-Coussa O., Obellianne M., Linderme D., Montes E., Maia-Grondard A., Vilaine F., Pannetier C. Wuschel overexpression promotes somatic embryogenesis and induces organogenesis in cotton (Gossypium hirsutum L.) tissues cultured in vitro. Plant Cell Rep. 2013;32:675–686. - PubMed
-
- Chen K., Wang Y., Zhang R., Zhang H., Gao C. CRISPR/Cas Genome Editing and Precision Plant Breeding in Agriculture. Annu. Rev. Plant Biol. 2019;70:667–697. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

