COV2Var, a function annotation database of SARS-CoV-2 genetic variation
- PMID: 37897356
- PMCID: PMC10767816
- DOI: 10.1093/nar/gkad958
COV2Var, a function annotation database of SARS-CoV-2 genetic variation
Abstract
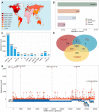
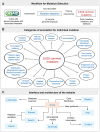
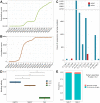
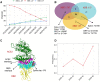
The COVID-19 pandemic, caused by the coronavirus SARS-CoV-2, has resulted in the loss of millions of lives and severe global economic consequences. Every time SARS-CoV-2 replicates, the viruses acquire new mutations in their genomes. Mutations in SARS-CoV-2 genomes led to increased transmissibility, severe disease outcomes, evasion of the immune response, changes in clinical manifestations and reducing the efficacy of vaccines or treatments. To date, the multiple resources provide lists of detected mutations without key functional annotations. There is a lack of research examining the relationship between mutations and various factors such as disease severity, pathogenicity, patient age, patient gender, cross-species transmission, viral immune escape, immune response level, viral transmission capability, viral evolution, host adaptability, viral protein structure, viral protein function, viral protein stability and concurrent mutations. Deep understanding the relationship between mutation sites and these factors is crucial for advancing our knowledge of SARS-CoV-2 and for developing effective responses. To fill this gap, we built COV2Var, a function annotation database of SARS-CoV-2 genetic variation, available at http://biomedbdc.wchscu.cn/COV2Var/. COV2Var aims to identify common mutations in SARS-CoV-2 variants and assess their effects, providing a valuable resource for intensive functional annotations of common mutations among SARS-CoV-2 variants.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
-
- Markov P.V., Ghafari M., Beer M., Lythgoe K., Simmonds P., Stilianakis N.I., Katzourakis A.. The evolution of SARS-CoV-2. Nat. Rev. Microbiol. 2023; 21:361–379. - PubMed
-
- Lauring A.S., Hodcroft E.B.. Genetic variants of SARS-CoV-2-what do they mean. JAMA. 2021; 325:529–531. - PubMed
-
- Xie X., Liu Y., Liu J., Zhang X., Zou J., Fontes-Garfias C.R., Xia H., Swanson K.A., Cutler M., Cooper D. Neutralization of SARS-CoV-2 spike 69/70 deletion, E484K and N501Y variants by BNT162b2 vaccine-elicited sera. Nat. Med. 2021; 27:620–621. - PubMed
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

