Structural and functional investigation of GajB protein in Gabija anti-phage defense
- PMID: 37897358
- PMCID: PMC10681800
- DOI: 10.1093/nar/gkad951
Structural and functional investigation of GajB protein in Gabija anti-phage defense
Abstract
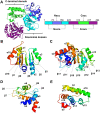
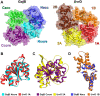
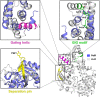
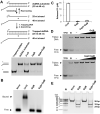
Bacteriophages (phages) are viruses that infect bacteria and archaea. To fend off invading phages, the hosts have evolved a variety of anti-phage defense mechanisms. Gabija is one of the most abundant prokaryotic antiviral systems and consists of two proteins, GajA and GajB. GajA has been characterized experimentally as a sequence-specific DNA endonuclease. Although GajB was previously predicted to be a UvrD-like helicase, its function is unclear. Here, we report the results of structural and functional analyses of GajB. The crystal structure of GajB revealed a UvrD-like domain architecture, including two RecA-like core and two accessory subdomains. However, local structural elements that are important for the helicase function of UvrD are not conserved in GajB. In functional assays, GajB did not unwind or bind various types of DNA substrates. We demonstrated that GajB interacts with GajA to form a heterooctameric Gabija complex, but GajB did not exhibit helicase activity when bound to GajA. These results advance our understanding of the molecular mechanism underlying Gabija anti-phage defense and highlight the role of GajB as a component of a multi-subunit antiviral complex in bacteria.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Ofir G., Sorek R.. Contemporary phage biology: from classic models to new insights. Cell. 2018; 172:1260–1270. - PubMed
-
- Fuhrman J.A. Marine viruses and their biogeochemical and ecological effects. Nature. 1999; 399:541–548. - PubMed
-
- Weinbauer M.G. Ecology of prokaryotic viruses. FEMS Microbiol. Rev. 2004; 28:127–181. - PubMed
-
- Hampton H.G., Watson B.N.J., Fineran P.C.. The arms race between bacteria and their phage foes. Nature. 2020; 577:327–336. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

