ADP-ribose contributions to genome stability and PARP enzyme trapping on sites of DNA damage; paradigm shifts for a coming-of-age modification
- PMID: 37898399
- PMCID: PMC10722394
- DOI: 10.1016/j.jbc.2023.105397
ADP-ribose contributions to genome stability and PARP enzyme trapping on sites of DNA damage; paradigm shifts for a coming-of-age modification
Abstract
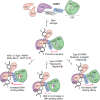
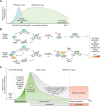
ADP-ribose is a versatile modification that plays a critical role in diverse cellular processes. The addition of this modification is catalyzed by ADP-ribosyltransferases, among which notable poly(ADP-ribose) polymerase (PARP) enzymes are intimately involved in the maintenance of genome integrity. The role of ADP-ribose modifications during DNA damage repair is of significant interest for the proper development of PARP inhibitors targeted toward the treatment of diseases caused by genomic instability. More specifically, inhibitors promoting PARP persistence on DNA lesions, termed PARP "trapping," is considered a desirable characteristic. In this review, we discuss key classes of proteins involved in ADP-ribose signaling (writers, readers, and erasers) with a focus on those involved in the maintenance of genome integrity. An overview of factors that modulate PARP1 and PARP2 persistence at sites of DNA lesions is also discussed. Finally, we clarify aspects of the PARP trapping model in light of recent studies that characterize the kinetics of PARP1 and PARP2 recruitment at sites of lesions. These findings suggest that PARP trapping could be considered as the continuous recruitment of PARP molecules to sites of lesions, rather than the physical stalling of molecules. Recent studies and novel research tools have elevated the level of understanding of ADP-ribosylation, marking a coming-of-age for this interesting modification.
Keywords: ADP-ribose; ADP-ribosyltransferase; ARH3; DNA damage; DNA strand breaks; PARG; PARP enzymes; PARP inhibitors; PARP trapping; allostery; chromatin; poly(ADP-ribose); poly(ADP-ribose) glycohydrolases; protein–nucleic acid interactions.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest J. M. P. is a cofounder of Hysplex LLC with interests in PARPi development. É. R.-T. declares no conflict of interest with the contents of this article.
Figures

References
-
- Lindahl T., Barnes D.E. Repair of endogenous DNA damage. Cold Spring Harb. Symp. Quant. Biol. 2000;65:127–133. - PubMed
-
- Huen M.S., Chen J. The DNA damage response pathways: at the crossroad of protein modifications. Cell Res. 2008;18:8–16. - PubMed
-
- Lüscher B., Bütepage M., Eckei L., Krieg S., Verheugd P., Shilton B.H. ADP-ribosylation, a multifaceted posttranslational modification involved in the control of cell physiology in health and disease. Chem. Rev. 2018;118:1092–1136. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

