A circular RNA activated by TGFβ promotes tumor metastasis through enhancing IGF2BP3-mediated PDPN mRNA stability
- PMID: 37898647
- PMCID: PMC10613289
- DOI: 10.1038/s41467-023-42571-1
A circular RNA activated by TGFβ promotes tumor metastasis through enhancing IGF2BP3-mediated PDPN mRNA stability
Abstract
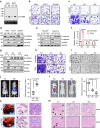
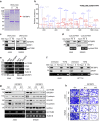
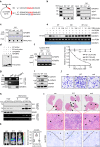
Metastasis is the leading cause of cancer-related death, where TGFβ-induced epithelial-mesenchymal transition (EMT) process confers on cancer cells increased metastatic potential. However, the involvement of circRNAs in this process is still obscure. Here, we identify a TGFβ-induced circRNA called circITGB6 as an indispensable factor during the TGFβ-mediated EMT process. circITGB6 is significantly upregulated in metastatic cancer samples and its higher abundance is closely correlated to worse prognosis of colorectal cancer (CRC) patients. Through gain- and loss-of-function assays, circITGB6 is found to potently promote EMT process and tumor metastasis in various models in vitro and in vivo. Mechanistically, circITGB6 enhances the mRNA stability of PDPN, an EMT-promoting gene, by directly interacting with IGF2BP3. Notably, interfering circITGB6 with PEI-coated specific siRNA effectively represses liver metastasis. Therefore, our study reveals the function of a TGFβ-regulated circRNA in tumor metastasis and suggests that targeting circITGB6 is a promising strategy for cancer therapy.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

