Genomic Mosaicism of the Brain: Origin, Impact, and Utility
- PMID: 37898991
- PMCID: PMC11178748
- DOI: 10.1007/s12264-023-01124-8
Genomic Mosaicism of the Brain: Origin, Impact, and Utility
Abstract
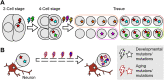
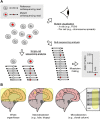
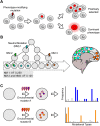
Genomic mosaicism describes the phenomenon where some but not all cells within a tissue harbor unique genetic mutations. Traditionally, research focused on the impact of genomic mosaicism on clinical phenotype-motivated by its involvement in cancers and overgrowth syndromes. More recently, we increasingly shifted towards the plethora of neutral mosaic variants that can act as recorders of cellular lineage and environmental exposures. Here, we summarize the current state of the field of genomic mosaicism research with a special emphasis on our current understanding of this phenomenon in brain development and homeostasis. Although the field of genomic mosaicism has a rich history, technological advances in the last decade have changed our approaches and greatly improved our knowledge. We will provide current definitions and an overview of contemporary detection approaches for genomic mosaicism. Finally, we will discuss the impact and utility of genomic mosaicism.
Keywords: Brain development; Brain homeostasis; Genomic mosaicism; Genomics.
© 2023. The Author(s).
Figures