Th17 cells target the metabolic miR-142-5p-succinate dehydrogenase subunit C/D (SDHC/SDHD) axis, promoting invasiveness and progression of cervical cancers
- PMID: 37899663
- PMCID: PMC11467798
- DOI: 10.1002/1878-0261.13546
Th17 cells target the metabolic miR-142-5p-succinate dehydrogenase subunit C/D (SDHC/SDHD) axis, promoting invasiveness and progression of cervical cancers
Abstract
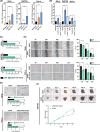
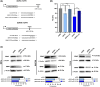
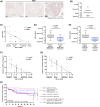
During cervical carcinogenesis, T-helper (Th)-17 cells accumulate in the peripheral blood and tumor tissues of cancer patients. We previously demonstrated that Th17 cells are associated with therapy resistance as well as cervical cancer metastases and relapse; however, the underlying Th17-driven mechanisms are not fully understood. Here, using microarrays, we found that Th17 cells induced an epithelial-to-mesenchymal transition (EMT) phenotype of cervical cancer cells and promoted migration and invasion of 2D cultures and 3D spheroids via induction of microRNA miR-142-5p. As the responsible mechanism, we identified the subunits C and D of the succinate dehydrogenase (SDH) complex as new targets of miR-142-5p and provided evidence that Th17-miR-142-5p-dependent reduced expression of SDHC and SDHD mediated enhanced migration and invasion of cancer cells using small interfering RNAs (siRNAs) for SDHC and SDHD, and miR-142-5p inhibitors. Consistently, patients exhibited high levels of succinate in their serum associated with lymph node metastases and diminished expression of SDHD in patient biopsies correlated with increased numbers of Th17 cells. Correspondingly, a combination of weak or negative SDHD expression and a ratio of Th17/CD4+ T cells > 43.90% in situ was associated with reduced recurrence-free survival. In summary, we unraveled a previously unknown molecular mechanism by which Th17 cells promote cervical cancer progression and suggest evaluation of Th17 cells as a potential target for immunotherapy in cervical cancer.
Keywords: T‐helper‐17 cells; cervical cancers; metastases; miR‐142‐5p; succinate dehydrogenase complex.
© 2023 The Authors. Molecular Oncology published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
LINC00319 promotes migration, invasion and epithelial-mesenchymal transition process in cervical cancer by regulating miR-3127-5p/RPP25 axis.In Vitro Cell Dev Biol Anim. 2020 Feb;56(2):145-153. doi: 10.1007/s11626-019-00425-5. Epub 2020 Jan 15. In Vitro Cell Dev Biol Anim. 2020. PMID: 31942724
-
MicroRNA-98-5p modulates cervical cancer progression via controlling PI3K/AKT pathway.Bioengineered. 2021 Dec;12(2):10596-10607. doi: 10.1080/21655979.2021.2000722. Bioengineered. 2021. PMID: 34895048 Free PMC article.
-
MicroRNA-361-5p facilitates cervical cancer progression through mediation of epithelial-to-mesenchymal transition.Med Oncol. 2013 Dec;30(4):751. doi: 10.1007/s12032-013-0751-0. Epub 2013 Oct 26. Med Oncol. 2013. PMID: 24158756
-
MiR-204-5p/TFAP2A feedback loop positively regulates the proliferation, migration, invasion and EMT process in cervical cancer.Cancer Biomark. 2020;28(3):381-390. doi: 10.3233/CBM-191064. Cancer Biomark. 2020. PMID: 32474464
-
LncRNA DANCR promotes cervical cancer progression by upregulating ROCK1 via sponging miR-335-5p.J Cell Physiol. 2019 May;234(5):7266-7278. doi: 10.1002/jcp.27484. Epub 2018 Oct 26. J Cell Physiol. 2019. PMID: 30362591
Cited by
-
Profiles of Circulating Exosomal miRNA in Patients with SAPHO Patients by High-Throughput Sequencing.Comb Chem High Throughput Screen. 2025;28(7):1193-1204. doi: 10.2174/0113862073289083240425114858. Comb Chem High Throughput Screen. 2025. PMID: 38752639
-
Identification of prognostic biomarkers for endometrioid endometrial carcinoma based on the miRNA and mRNA co-expression network regulated by estradiol.Clinics (Sao Paulo). 2025 May 3;80:100672. doi: 10.1016/j.clinsp.2025.100672. eCollection 2025. Clinics (Sao Paulo). 2025. PMID: 40319860 Free PMC article.
References
-
- Heeren AM, Koster BD, Samuels S, Ferns DM, Chondronasiou D, Kenter GG, et al. High and interrelated rates of PD‐L1+CD14+ antigen‐presenting cells and regulatory T cells mark the microenvironment of metastatic lymph nodes from patients with cervical cancer. Cancer Immunol Res. 2015;3:48–58. 10.1158/2326-6066.CIR-14-0149 - DOI - PubMed

