The role of the plasmid-mediated fluoroquinolone resistance genes as resistance mechanisms in pediatric infections due to Enterobacterales
- PMID: 37900312
- PMCID: PMC10613066
- DOI: 10.3389/fcimb.2023.1249505
The role of the plasmid-mediated fluoroquinolone resistance genes as resistance mechanisms in pediatric infections due to Enterobacterales
Abstract
Introduction: Fluoroquinolones (FQs) are not commonly prescribed in children, yet the increasing incidence of multidrug-resistant (MDR) Enterobacterales (Ent) infections in this population often reveals FQ resistance. We sought to define the role of FQ resistance in the epidemiology of MDR Ent in children, with an overall goal to devise treatment and prevention strategies.
Methods: A case-control study of children (0-18 years) at three Chicago hospitals was performed. Cases had infections by FQ-susceptible, β-lactamase-producing (bla) Ent harboring a non- or low-level expression of PMFQR genes (PMFQS Ent). Controls had FQR infections due to bla Ent with expressed PMFQR genes (PMFQR Ent). We sought bla genes by PCR or DNA (BD Max Check-Points assay®) and PMFQR genes by PCR. We performed rep-PCR, MLST, and E. coli phylogenetic grouping. Whole genome sequencing was additionally performed on PMFQS Ent positive isolates. Demographics, comorbidities, and device, antibiotic, and healthcare exposures were evaluated. Predictors of infection were assessed.
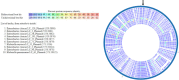
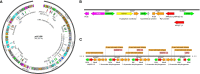
Results: Of 170 β-lactamase-producing Ent isolates, 85 (50%) were FQS; 23 (27%) had PMFQR genes (PMFQS cases). Eighty-five (50%) were FQR; 53 (62%) had PMFQR genes (PMFQR controls). The median age for children with PMFQS Ent and PMFQR Ent was 4.3 and 6.2 years, respectively (p = NS). Of 23 PMFQS Ent, 56% were Klebsiella spp., and of 53 PMFQR Ent, 76% were E. coli. The most common bla and PMFQR genes detected in PMFQS Ent were bla SHV ESBL (44%) and oqxAB (57%), and the corresponding genes detected in PMFQR Ent were bla CTX-M-1-group ESBL (79%) and aac(6')-Ib-cr (83%). Whole genome sequencing of PMFQS Ent revealed the additional presence of mcr-9, a transferable polymyxin resistance gene, in 47% of isolates, along with multiple plasmids and mobile genetic elements propagating drug resistance. Multivariable regression analysis showed that children with PMFQS Ent infections were more likely to have hospital onset infection (OR 5.7, 95% CI 1.6-22) and isolates containing multiple bla genes (OR 3.8, 95% CI 1.1-14.5). The presence of invasive devices mediated the effects of healthcare setting in the final model. Differences in demographics, comorbidities, or antibiotic use were not found.
Conclusions: Paradoxically, PMFQS Ent infections were often hospital onset and PMFQR Ent infections were community onset. PMFQS Ent commonly co-harbored multiple bla and PMFQR genes, and additional silent, yet transferrable antibiotic resistance genes such as mcr-9, affecting therapeutic options and suggesting the need to address infection prevention strategies to control spread. Control of PMFQS Ent infections will require validating community and healthcare-based sources and risk factors associated with acquisition.
Keywords: Enterobacterales infections; antibiotic resistance; beta-lactamases; children; epidemiology; fluoroquinolone resistance; gram-negative bacteria; mcr-9.
Copyright © 2023 Logan, Coy, Pitstick, Marshall, Medernach, Domitrovic, Konda, Qureshi, Hujer, Zheng, Rudin, Weinstein and Bonomo.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- Bingen-Bidois M., Clermont O., Bonacorsi S., Terki M., Brahimi N., Loukil C., et al. . (2002). Phylogenetic analysis and prevalence of urosepsis strains of Escherichia coli bearing pathogenicity island-like domains. Infect. Immun. 70 (6), 3216–3226. doi: 10.1128/IAI.70.6.3216-3226.2002 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

