Assessment of Donor Derived Cell Free DNA (dd-cfDNA) at Surveillance and at Clinical Suspicion of Acute Rejection in Renal Transplantation
- PMID: 37901296
- PMCID: PMC10603235
- DOI: 10.3389/ti.2023.11507
Assessment of Donor Derived Cell Free DNA (dd-cfDNA) at Surveillance and at Clinical Suspicion of Acute Rejection in Renal Transplantation
Abstract
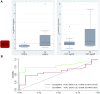
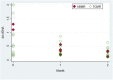
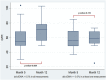
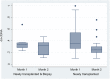
In our prospective, unicenter cohort study, we collected blood samples from 30 newly kidney transplanted patients, at month 1, 2, 3, and 5 for dd-cfDNA analysis, along with creatinine/eGFR and DSA monitoring, and from 32 patients who underwent an indication biopsy and whose dd-cfDNA levels were measured at the time of biopsy and 1 month afterwards. Fourteen of 32 (43.8%) patients in the biopsy group were diagnosed with TCMR and 5 of 32 (15.6%) with ABMR. Dd-cfDNA proved to be better than creatinine in diagnosing rejection from non-rejection in patients who were biopsied. When a dd-cfDNA threshold of 0.5% was chosen, sensitivity was 73.7% and specificity was 92.3% (AUC: 0.804, 0.646-0.961). In rejection patients, levels of dd-cfDNA prior to biopsy (0.94%, 0.3-2.0) decreased substantially after initiation of treatment with median returning to baseline already at 1 month (0.33%, 0.21-0.51, p = 0.0036). In the surveillance group, high levels of dd-cfDNA (>0.5%) from second month post-transplantation were correlated with non-increasing eGFR 1 year post-transplantation. The study used AlloSeq kit for kidney transplant surveillance for first time and confirmed dd-cfDNA's ability to detect rejection and monitor treatment, as well as to predict worse long-term outcomes regarding eGFR.
Keywords: biomarker; dd-cfDNA; kidney allograft; rejection; transplantation.
Copyright © 2023 Mantios, Filiopoulos, Constantoulakis, Liapis, Vittoraki, Casas, Marinaki and Boletis.
Conflict of interest statement
The study received a grant from CareDx and Genotypos-Bioanalytica. SC has an affiliation with CareDx. PC has an affiliation with Genotypos-Bioanalytica. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Comment in
-
Donor-Derived Cell-Free DNA: Attractive Biomarker Seeks a Context of Use.Transpl Int. 2023 Dec 1;36:12406. doi: 10.3389/ti.2023.12406. eCollection 2023. Transpl Int. 2023. PMID: 38106814 Free PMC article. No abstract available.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

