Isolation and identification of antagonistic Bacillus amyloliquefaciens HSE-12 and its effects on peanut growth and rhizosphere microbial community
- PMID: 37901825
- PMCID: PMC10601714
- DOI: 10.3389/fmicb.2023.1274346
Isolation and identification of antagonistic Bacillus amyloliquefaciens HSE-12 and its effects on peanut growth and rhizosphere microbial community
Abstract
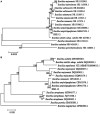
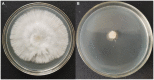
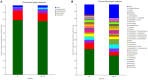
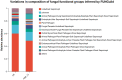
The HSE-12 strain isolated from peanut rhizosphere soil was identified as Bacillus amyloliquefaciens by observation of phenotypic characteristics, physiological and biochemical tests, 16S rDNA and gyrB gene sequencing. In vitro experiments showed that the strain possessed biocontrol activity against a variety of pathogens including Sclerotium rolfsii. The strain has the ability to produce hydrolytic enzymes, as well as volatile organic compounds with antagonistic and probiotic effects such as ethyleneglycol and 2,3-butanediol. In addition, HSE-12 showed potassium solubilizing (10.54 ± 0.19 mg/L), phosphorus solubilization (168.34 ± 8.06 mg/L) and nitrogen fixation (17.35 ± 2.34 mg/g) abilities, and was able to secrete siderophores [(Ar-A)/Ar × 100%: 56%] which promoted plant growth. After inoculating peanut with HSE-12, the available phosphorus content in rhizosphere soil increased by 27%, urease activity increased by 43%, catalase activity increased by 70% and sucrase activity increased by 50% (p < 0.05). The dry weight, fresh weight and the height of the first pair of lateral branches of peanuts increased by 24.7, 41.9, and 36.4%, respectively, compared with uninoculated peanuts. In addition, compared with the blank control, it increased the diversity and richness of peanut rhizosphere bacteria and changed the community structure of bacteria and fungi. The relative abundance of beneficial microorganisms such as Sphingomonas, Arthrobacter, RB41, and Micromonospora in rhizosphere soil was increased, while the relative abundance of pathogenic microorganisms such as Aspergillus, Neocosmospora, and Rhizoctonia was decreased.
Keywords: Bacillus amyloliquefaciens; biocontrol; microbial community; plant growth-promoting rhizobacteria; volatile compounds.
Copyright © 2023 Li, Li, Song, Li, Zhang, Sun, Geng and Liu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Evaluation of efficacy and mechanism of Bacillus velezensis CB13 for controlling peanut stem rot caused by Sclerotium rolfsii.Front Microbiol. 2023 Feb 16;14:1111965. doi: 10.3389/fmicb.2023.1111965. eCollection 2023. Front Microbiol. 2023. PMID: 36876084 Free PMC article.
-
Plant Growth-Promoting Rhizobacteria Promote Growth of Seedlings, Regulate Soil Microbial Community, and Alleviate Damping-Off Disease Caused by Rhizoctonia solani on Pinus sylvestris var. mongolica.Plant Dis. 2022 Oct;106(10):2730-2740. doi: 10.1094/PDIS-11-21-2562-RE. Epub 2022 Sep 12. Plant Dis. 2022. PMID: 36094426
-
Plant-Microbe Interaction: Mining the Impact of Native Bacillus amyloliquefaciens WS-10 on Tobacco Bacterial Wilt Disease and Rhizosphere Microbial Communities.Microbiol Spectr. 2022 Aug 31;10(4):e0147122. doi: 10.1128/spectrum.01471-22. Epub 2022 Aug 1. Microbiol Spectr. 2022. PMID: 35913211 Free PMC article.
-
Identification of a phosphorus-solubilizing Tsukamurella tyrosinosolvens strain and its effect on the bacterial diversity of the rhizosphere soil of peanuts growth-promoting.World J Microbiol Biotechnol. 2021 May 31;37(7):109. doi: 10.1007/s11274-021-03078-3. World J Microbiol Biotechnol. 2021. PMID: 34057641
-
Isolation and Genome Sequence of a Novel Phosphate-Solubilizing Rhizobacterium Bacillus altitudinis GQYP101 and Its Effects on Rhizosphere Microbial Community Structure and Functional Traits of Corn Seedling.Curr Microbiol. 2022 Jul 14;79(9):249. doi: 10.1007/s00284-022-02944-z. Curr Microbiol. 2022. PMID: 35834051 Review.
Cited by
-
Characterization of potassium-solubilizing fungi, Mortierella spp., isolated from a poplar plantation rhizosphere soil.Arch Microbiol. 2024 Mar 13;206(4):157. doi: 10.1007/s00203-024-03912-w. Arch Microbiol. 2024. PMID: 38480543
-
Phosphate-Solubilizing Bacteria: Advances in Their Physiology, Molecular Mechanisms and Microbial Community Effects.Microorganisms. 2023 Dec 1;11(12):2904. doi: 10.3390/microorganisms11122904. Microorganisms. 2023. PMID: 38138048 Free PMC article. Review.
-
Metagenomic Analysis to Assess the Impact of Plant Growth-Promoting Rhizobacteria on Peanut (Arachis hypogaea L.) Crop Production and Soil Enzymes and Microbial Diversity.J Agric Food Chem. 2024 Oct 9;72(40):22385-22397. doi: 10.1021/acs.jafc.4c05687. Epub 2024 Sep 26. J Agric Food Chem. 2024. PMID: 39324627 Free PMC article.
-
Isolation of Bacillus amyloliquefaciens D39 and Identification of Its Antimicrobial Proteins Active Against Chestnut Blight.Microorganisms. 2025 Jun 3;13(6):1302. doi: 10.3390/microorganisms13061302. Microorganisms. 2025. PMID: 40572190 Free PMC article.
-
Enhancing carrot (Daucus carota var. sativa Hoffm.) plant productivity with combined rhizosphere microbial consortium.Front Microbiol. 2024 Nov 20;15:1466300. doi: 10.3389/fmicb.2024.1466300. eCollection 2024. Front Microbiol. 2024. PMID: 39633805 Free PMC article.
References
-
- Bao S. (2013). Soil and agricultural chemistry analysis. Beijing, China: Agriculture Press Publisher.
-
- Chen C., Bauske E. M., Musson G., Rodriguezkabana R., Kloepper J. W. (1995). Biological control of fusarium wilt on cotton by use of endophytic bacteria. Biol. Control 5, 83–91. doi: 10.1006/bcon.1995.1009 - DOI
-
- Cui Y. T., Lv Y., Song M. Q., Wang S., Hu H. T., Jahan N. S., et al. (2020). Genome sequence of Micromonospora terminaliae TMS7(T), a new endophytic actinobacterium isolated from the medicinal plant Terminalia mucronata. Mol. Plant-Microbe Interact. 33, 721–723. doi: 10.1094/mpmi-12-19-0336-a, PMID: - DOI - PubMed
-
- Dame Z. T., Rahman M., Islam T. (2021). Bacilli as sources of agrobiotechnology: recent advances and future directions. Green Chem. Lett. Rev. 14, 246–271. doi: 10.1080/17518253.2021.1905080 - DOI
LinkOut - more resources
Full Text Sources
Research Materials

