Aberrant DJ-1 expression underlies L-type calcium channel hypoactivity in dendrites in tuberous sclerosis complex and Alzheimer's disease
- PMID: 37903257
- PMCID: PMC10636362
- DOI: 10.1073/pnas.2301534120
Aberrant DJ-1 expression underlies L-type calcium channel hypoactivity in dendrites in tuberous sclerosis complex and Alzheimer's disease
Abstract
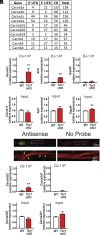
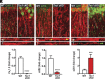
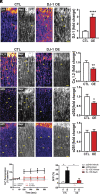
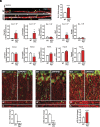
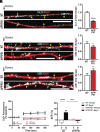
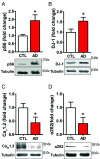
L-type voltage-gated calcium (Ca2+) channels (L-VGCC) dysfunction is implicated in several neurological and psychiatric diseases. While a popular therapeutic target, it is unknown whether molecular mechanisms leading to disrupted L-VGCC across neurodegenerative disorders are conserved. Importantly, L-VGCC integrate synaptic signals to facilitate a plethora of cellular mechanisms; however, mechanisms that regulate L-VGCC channel density and subcellular compartmentalization are understudied. Herein, we report that in disease models with overactive mammalian target of rapamycin complex 1 (mTORC1) signaling (or mTORopathies), deficits in dendritic L-VGCC activity are associated with increased expression of the RNA-binding protein (RBP) Parkinsonism-associated deglycase (DJ-1). DJ-1 binds the mRNA coding for the alpha and auxiliary Ca2+ channel subunits CaV1.2 and α2δ2, and represses their mRNA translation, only in the disease states, specifically preclinical models of tuberous sclerosis complex (TSC) and Alzheimer's disease (AD). In agreement, DJ-1-mediated repression of CaV1.2/α2δ2 protein synthesis in dendrites is exaggerated in mouse models of AD and TSC, resulting in deficits in dendritic L-VGCC calcium activity. Finding of DJ-1-regulated L-VGCC activity in dendrites in TSC and AD provides a unique signaling pathway that can be targeted in clinical mTORopathies.
Keywords: Alzheimer’s disease; RNA-binding protein; mammalian target of rapamycin; tuberous sclerosis complex; voltage-gated calcium channels.
Conflict of interest statement
The authors declare no competing interest.
Figures

References
-
- Modi M. E., Sahin M., Tau: A novel entry point for mTOR-based treatments in autism spectrum disorder? Neuron 106, 359–361 (2020). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AG073823/AG/NIA NIH HHS/United States
- R01 AG055581/AG/NIA NIH HHS/United States
- R01 AA029691/AA/NIAAA NIH HHS/United States
- T32 AA007565/AA/NIAAA NIH HHS/United States
- R01 NS105005/NS/NINDS NIH HHS/United States
- R01 AA026551/AA/NIAAA NIH HHS/United States
- R01 AG056622/AG/NIA NIH HHS/United States
- P30 AG066509/AG/NIA NIH HHS/United States
- R21 AG077271/AG/NIA NIH HHS/United States
- R16 NS134542/NS/NINDS NIH HHS/United States
- T32 DA041349/DA/NIDA NIH HHS/United States
- P30 AG072947/AG/NIA NIH HHS/United States
- U19 AG066567/AG/NIA NIH HHS/United States
- P30 AG049638/AG/NIA NIH HHS/United States
- F31 NS117096/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

