PhysiBoSS 2.0: a sustainable integration of stochastic Boolean and agent-based modelling frameworks
- PMID: 37903760
- PMCID: PMC10616087
- DOI: 10.1038/s41540-023-00314-4
PhysiBoSS 2.0: a sustainable integration of stochastic Boolean and agent-based modelling frameworks
Abstract
In systems biology, mathematical models and simulations play a crucial role in understanding complex biological systems. Different modelling frameworks are employed depending on the nature and scales of the system under study. For instance, signalling and regulatory networks can be simulated using Boolean modelling, whereas multicellular systems can be studied using agent-based modelling. Herein, we present PhysiBoSS 2.0, a hybrid agent-based modelling framework that allows simulating signalling and regulatory networks within individual cell agents. PhysiBoSS 2.0 is a redesign and reimplementation of PhysiBoSS 1.0 and was conceived as an add-on that expands the PhysiCell functionalities by enabling the simulation of intracellular cell signalling using MaBoSS while keeping a decoupled, maintainable and model-agnostic design. PhysiBoSS 2.0 also expands the set of functionalities offered to the users, including custom models and cell specifications, mechanistic submodels of substrate internalisation and detailed control over simulation parameters. Together with PhysiBoSS 2.0, we introduce PCTK, a Python package developed for handling and processing simulation outputs, and generating summary plots and 3D renders. PhysiBoSS 2.0 allows studying the interplay between the microenvironment, the signalling pathways that control cellular processes and population dynamics, suitable for modelling cancer. We show different approaches for integrating Boolean networks into multi-scale simulations using strategies to study the drug effects and synergies in models of cancer cell lines and validate them using experimental data. PhysiBoSS 2.0 is open-source and publicly available on GitHub with several repositories of accompanying interoperable tools.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

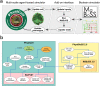


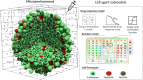



References
-
- Kitano, H. Foundations of Systems Biology (The MIT Press, 2001).
Publication types
MeSH terms
Grants and funding
- 951773/EC | Horizon 2020 Framework Programme (EU Framework Programme for Research and Innovation H2020)
- 101092749/EC | Horizon 2020 Framework Programme (EU Framework Programme for Research and Innovation H2020)
- 825070/EC | Horizon 2020 Framework Programme (EU Framework Programme for Research and Innovation H2020)
- 951773/EC | Horizon 2020 Framework Programme (EU Framework Programme for Research and Innovation H2020)
- 101092749/EC | Horizon 2020 Framework Programme (EU Framework Programme for Research and Innovation H2020)
- 825070/EC | Horizon 2020 Framework Programme (EU Framework Programme for Research and Innovation H2020)
- 951773/EC | Horizon 2020 Framework Programme (EU Framework Programme for Research and Innovation H2020)
- 101092749/EC | Horizon 2020 Framework Programme (EU Framework Programme for Research and Innovation H2020)
- 951773/EC | Horizon 2020 Framework Programme (EU Framework Programme for Research and Innovation H2020)
- 825070/EC | Horizon 2020 Framework Programme (EU Framework Programme for Research and Innovation H2020)
- 825070/EC | Horizon 2020 Framework Programme (EU Framework Programme for Research and Innovation H2020)
- 951773/EC | Horizon 2020 Framework Programme (EU Framework Programme for Research and Innovation H2020)
- ANR-19-P3IA-0001/Agence Nationale de la Recherche (French National Research Agency)
- 951773/EC | Horizon 2020 Framework Programme (EU Framework Programme for Research and Innovation H2020)
LinkOut - more resources
Full Text Sources
Medical

