YTHDC1 as a tumor progression suppressor through modulating FSP1-dependent ferroptosis suppression in lung cancer
- PMID: 37903990
- PMCID: PMC10733405
- DOI: 10.1038/s41418-023-01234-w
YTHDC1 as a tumor progression suppressor through modulating FSP1-dependent ferroptosis suppression in lung cancer
Abstract
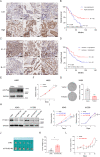
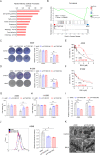
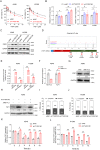
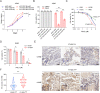
Ferroptosis is a regulated cell death process initiated by iron-dependent phospholipid peroxidation and is mainly suppressed by GPX4-dependent and FSP1-dependent surveillance mechanisms. However, how the ferroptosis surveillance system is regulated during cancer development remains largely unknown. Here, we report that the YTHDC1-mediated m6A epigenetic regulation of FSP1 alleviates the FSP1-dependent ferroptosis suppression that partially contributes to the tumor suppressive role of YTHDC1 in lung cancer progression. YTHDC1 knockdown promoted the lung tumor progression and upregulated FSP1 protein level that resulted in ferroptosis resistance of lung cancer cells. Silencing FSP1 abrogated YTHDC1 knockdown-induced proliferation increase and ferroptosis resistance. Mechanistically, YTHDC1 binding to the m6A sites in the FSP1 3'-UTR recruited the alternative polyadenylation regulator CSTF3 to generate a less stable shorter 3'-UTR contained FSP1 mRNA, whereas YTHDC1 downregulation generated the longer 3'-UTR contained FSP1 mRNA that is stabilized by RNA binding protein HuR and thus led to the enhanced FSP1 protein level. Therefore, our findings identify YTHDC1 as a tumor progression suppressor in lung cancer and a ferroptosis regulator through modulating the FSP1 mRNA stability and thus suggest a ferroptosis-related therapeutic option for YTHDC1high lung cancer.
© 2023. The Author(s), under exclusive licence to ADMC Associazione Differenziamento e Morte Cellulare.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Wu M, Xu LG, Su T, Tian Y, Zhai Z, Shu HB. AMID is a p53-inducible gene downregulated in tumors. Oncogene. 2004;23:6815–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

