Taxonomic update for giant viruses in the order Imitervirales (phylum Nucleocytoviricota)
- PMID: 37904060
- PMCID: PMC11230039
- DOI: 10.1007/s00705-023-05906-3
Taxonomic update for giant viruses in the order Imitervirales (phylum Nucleocytoviricota)
Abstract
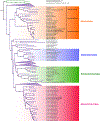
Large DNA viruses in the phylum Nucleocytoviricota, sometimes referred to as "giant viruses" owing to their large genomes and virions, have been the subject of burgeoning interest over the last decade. Here, we describe recently adopted taxonomic updates for giant viruses within the order Imitervirales. The families Allomimiviridae, Mesomimiviridae, and Schizomimiviridae have been created to accommodate the increasing diversity of mimivirus relatives that have sometimes been referred to in the literature as "extended Mimiviridae". In addition, the subfamilies Aliimimivirinae, Megamimivirinae, and Klosneuvirinae have been established to refer to subgroups of the Mimiviridae. Binomial names have also been adopted for all recognized species in the order. For example, Acanthamoeba polyphaga mimivirus is now classified in the species Mimivirus bradfordmassiliense.
© 2023. The Author(s), under exclusive licence to Springer-Verlag GmbH Austria, part of Springer Nature.
Conflict of interest statement
Declarations
Figures
References
-
- Abergel C, Legendre M, Claverie J-M (2015) The rapidly expanding universe of giant viruses: Mimivirus, Pandoravirus, Pithovirus and Mollivirus. FEMS Microbiol Rev 39:779–796 - PubMed
-
- Endo H, Blanc-Mathieu R, Li Y et al. (2020) Biogeography of marine giant viruses reveals their interplay with eukaryotes and ecological functions. Nat Ecol Evol 4:1639–1649 - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

