Chemical Expansion of the Methyltransferase Reaction: Tools for DNA Labeling and Epigenome Analysis
- PMID: 37904501
- PMCID: PMC10666283
- DOI: 10.1021/acs.accounts.3c00471
Chemical Expansion of the Methyltransferase Reaction: Tools for DNA Labeling and Epigenome Analysis
Abstract
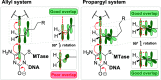
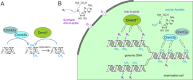
DNA is the genetic matter of life composed of four major nucleotides which can be further furnished with biologically important covalent modifications. Among the variety of enzymes involved in DNA metabolism, AdoMet-dependent methyltransferases (MTases) combine the recognition of specific sequences and covalent methylation of a target nucleotide. The naturally transferred methyl groups play important roles in biological signaling, but they are poor physical reporters and largely resistant to chemical derivatization. Therefore, an obvious strategy to unlock the practical utility of the methyltransferase reactions is to enable the transfer of "prederivatized" (extended) versions of the methyl group.However, previous enzymatic studies of extended AdoMet analogs indicated that the transalkylation reactions are drastically impaired as the size of the carbon chain increases. In collaborative efforts, we proposed that, akin to enhanced SN2 reactivity of allylic and propargylic systems, addition of a π orbital next to the transferable carbon atom might confer the needed activation of the reaction. Indeed, we found that MTase-catalyzed transalkylations of DNA with cofactors containing a double or a triple C-C bond in the β position occurred in a robust and sequence-specific manner. Altogether, this breakthrough approach named mTAG (methyltransferase-directed transfer of activated groups) has proven instrumental for targeted labeling of DNA and other types of biomolecules (using appropriate MTases) including RNA and proteins.Our further work focused on the propargylic cofactors and their reactions with DNA cytosine-5 MTases, a class of MTases common for both prokaryotes and eukaryotes. Here, we learned that the 4-X-but-2-yn-1-yl (X = polar group) cofactors suffered from a rapid loss of activity in aqueous buffers due to susceptibility of the triple bond to hydration. This problem was remedied by synthetically increasing the separation between X and the triple bond from one to three carbon units (6-X-hex-2-ynyl cofactors). To further optimize the transfer of the bulkier groups, we performed structure-guided engineering of the MTase cofactor pocket. Alanine replacements of two conserved residues conferred substantial improvements of the transalkylation activity with M.HhaI and three other engineered bacterial C5-MTases. Of particular interest were CpG-specific DNA MTases (M.SssI), which proved valuable tools for studies of mammalian methylomes and chemical probing of DNA function.Inspired by the successful repurposing of bacterial enzymes, we turned to more complex mammalian C5-MTases (Dnmt1, Dnmt3A, and Dnmt3B) and asked if they could ultimately lead to mTAG labeling inside mammalian cells. Our efforts to engineer mouse Dnmt1 produced a variant (Dnmt1*) that enabled efficient Dnmt1-directed deposition of 6-azide-hexynyl groups on DNA in vitro. CRISPR-Cas9 editing of the corresponding codons in the genomic Dnmt1 alleles established endogenous expression of Dnmt1* in mouse embryonic stem cells. To circumvent the poor cellular uptake of AdoMet and its analogs, we elaborated their efficient internalization by electroporation, which has finally enabled selective catalysis-dependent azide tagging of natural Dnmt1 targets in live mammalian cells. The deposited chemical groups were then exploited as "click" handles for reading adjoining sequences and precise genomic mapping of the methylation sites. These findings offer unprecedented inroads into studies of DNA methylation in a wide range of eukaryotic model systems.
Conflict of interest statement
The authors declare the following competing financial interest(s): The authors are inventors on several patents related to this work.
Figures


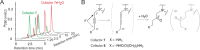


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

