DisProt in 2024: improving function annotation of intrinsically disordered proteins
- PMID: 37904585
- PMCID: PMC10767923
- DOI: 10.1093/nar/gkad928
DisProt in 2024: improving function annotation of intrinsically disordered proteins
Erratum in
-
Correction to 'DisProt in 2024: improving function annotation of intrinsically disordered proteins'.Nucleic Acids Res. 2025 Feb 27;53(5):gkaf228. doi: 10.1093/nar/gkaf228. Nucleic Acids Res. 2025. PMID: 40087889 Free PMC article. No abstract available.
Abstract
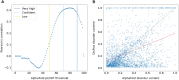
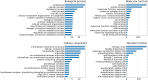
DisProt (URL: https://disprot.org) is the gold standard database for intrinsically disordered proteins and regions, providing valuable information about their functions. The latest version of DisProt brings significant advancements, including a broader representation of functions and an enhanced curation process. These improvements aim to increase both the quality of annotations and their coverage at the sequence level. Higher coverage has been achieved by adopting additional evidence codes. Quality of annotations has been improved by systematically applying Minimum Information About Disorder Experiments (MIADE) principles and reporting all the details of the experimental setup that could potentially influence the structural state of a protein. The DisProt database now includes new thematic datasets and has expanded the adoption of Gene Ontology terms, resulting in an extensive functional repertoire which is automatically propagated to UniProtKB. Finally, we show that DisProt's curated annotations strongly correlate with disorder predictions inferred from AlphaFold2 pLDDT (predicted Local Distance Difference Test) confidence scores. This comparison highlights the utility of DisProt in explaining apparent uncertainty of certain well-defined predicted structures, which often correspond to folding-upon-binding fragments. Overall, DisProt serves as a comprehensive resource, combining experimental evidence of disorder information to enhance our understanding of intrinsically disordered proteins and their functional implications.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
-
- Tompa P., Fersht A.. Structure and Function of Intrinsically Disordered Proteins. 2009; CRC Press.
-
- Ruan H., Sun Q., Zhang W., Liu Y., Lai L.. Targeting intrinsically disordered proteins at the edge of chaos. Drug Discov. Today. 2019; 24:217–227. - PubMed

