SUCLG2 Regulates Mitochondrial Dysfunction through Succinylation in Lung Adenocarcinoma
- PMID: 37904651
- PMCID: PMC10724390
- DOI: 10.1002/advs.202303535
SUCLG2 Regulates Mitochondrial Dysfunction through Succinylation in Lung Adenocarcinoma
Abstract
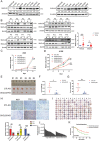
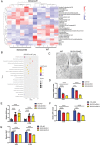
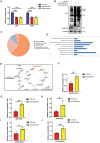
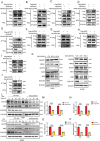
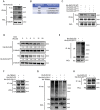
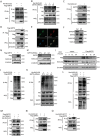
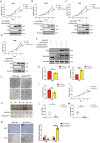
Mitochondrial dysfunction and abnormal energy metabolism are major features of cancer. However, the mechanisms underlying mitochondrial dysfunction during cancer progression are far from being clarified. Here, it is demonstrated that the expression level of succinyl-coenzyme A (CoA) synthetase GDP-forming subunit β (SUCLG2) can affect the overall succinylation of lung adenocarcinoma (LUAD) cells. Succinylome analysis shows that the deletion of SUCLG2 can upregulate the succinylation level of mitochondrial proteins and inhibits the function of key metabolic enzymes by reducing either enzymatic activity or protein stability, thus dampening mitochondrial function in LUAD cells. Interestingly, SUCLG2 itself is also succinylated on Lys93, and this succinylation enhances its protein stability, leading to the upregulation of SUCLG2 and promoting the proliferation and tumorigenesis of LUAD cells. Sirtuin 5 (SIRT5) desuccinylates SUCLG2 on Lys93, followed by tripartite motif-containing protein 21 (TRIM21)-mediated ubiquitination through K63-linkage and degradation in the lysosome. The findings reveal a new role for SUCLG2 in mitochondrial dysfunction and clarify the mechanism of the succinylation-mediated protein homeostasis of SUCLG2 in LUAD, thus providing a theoretical basis for developing anti-cancer drugs targeting SUCLG2.
Keywords: SIRT5; SUCLG2; TRIM21; mitochondrial dysfunction; succinylation.
© 2023 The Authors. Advanced Science published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
-
- Hanahan D., Cancer Discov. 2022, 12, 31. - PubMed
-
- Miller C., Wang L., Ostergaard E., Dan P., Saada A., Biochim. Biophys. Acta 2011, 1812, 625. - PubMed
-
- Lin S. R., Wen Y. C., Yeh H. L., Jiang K. C., Chen W. H., Mokgautsi N., Huang J., Chen W. Y., Liu Y. N., Oncogene 2020, 39, 6757. - PubMed
-
- Hadrava Vanova K., Pang Y., Krobova L., Kraus M., Nahacka Z., Boukalova S., Pack S. D., Zobalova R., Zhu J., Huynh T. T., Jochmanova I., Uher O., Hubackova S., Dvorakova S., Garrett T. J., Ghayee H. K., Wu X., Schuster B., Knapp P. E., Frysak Z., Hartmann I., Nilubol N., Cerny J., Taieb D., Rohlena J., Neuzil J., Yang C., Pacak K., J. Natl. Cancer Inst. 2022, 114, 130. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
