Conformational features and ionization states of Lys side chains in a protein studied using the stereo-array isotope labeling (SAIL) method
- PMID: 37904773
- PMCID: PMC10539808
- DOI: 10.5194/mr-2-223-2021
Conformational features and ionization states of Lys side chains in a protein studied using the stereo-array isotope labeling (SAIL) method
Abstract
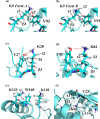
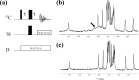
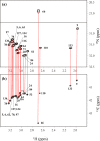
Although both the hydrophobic aliphatic chain and hydrophilic -amino group of the Lys side chain presumably contribute to the structures and functions of proteins, the dual nature of the Lys residue has not been fully investigated using NMR spectroscopy, due to the lack of appropriate methods to acquire comprehensive information on its long consecutive methylene chain. We describe herein a robust strategy to address the current situation, using various isotope-aided NMR technologies. The feasibility of our approach is demonstrated for the PHS/V66K variant of staphylococcal nuclease (SNase), which contains 21 Lys residues, including the engineered Lys-66 with an unusually low p of 5.6. All of the NMR signals for the 21 Lys residues were sequentially and stereospecifically assigned using the stereo-array isotope-labeled Lys (SAIL-Lys), [U-C,N; ,,,-D]-Lys. The complete set of assigned H, C, and N NMR signals for the Lys side-chain moieties affords useful structural information. For example, the set includes the characteristic chemical shifts for the C, C, and N signals for Lys-66, which has the deprotonated -amino group, and the large upfield shifts for the H and C signals for the Lys-9, Lys-28, Lys-84, Lys-110, and Lys-133 side chains, which are indicative of nearby aromatic rings. The C and N chemical shifts of the SNase variant selectively labeled with either [-C;,-D]-Lys or SAIL-Lys, dissolved in HO and DO, showed that the deuterium-induced shifts for Lys-66 were substantially different from those of the other 20 Lys residues. Namely, the deuterium-induced shifts of the C and N signals depend on the ionization states of the -amino group, i.e., 0.32 ppm for C [ND-NH] vs. 0.21 ppm for C [ND-NH] and 1.1 ppm for N[ND-NH] vs. 1.8 ppm for N[ND-NH]. Since the 1D C NMR spectrum of a protein selectively labeled with [-C;,-D]-Lys shows narrow ( 2 Hz) and well-dispersed C signals, the deuterium-induced shift difference of 0.11 ppm for the protonated and deprotonated -amino groups, which corresponds to 16.5 Hz at a field strength of 14 T (150 MHz for C), could be accurately measured. Although the isotope shift difference itself may not be absolutely decisive to distinguish the ionization state of the -amino group, the C, C, and N signals for a Lys residue with a deprotonated -amino group are likely to exhibit distinctive chemical shifts as compared to the normal residues with protonated -amino groups. Therefore, the isotope shifts would provide a useful auxiliary index for identifying Lys residues with deprotonated -amino groups at physiological pH levels.
Copyright: © 2021 Mitsuhiro Takeda et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures






References
-
- Cavanagh J, Fairbrother WJ, Palmer AG, Rance M, Skelton JJ. Protein NMR Spectroscopy: Principles and Practice. Academic Press; New York, USA: 2007.
LinkOut - more resources
Full Text Sources
