This is a preprint.
Immune landscape of tertiary lymphoid structures in hepatocellular carcinoma (HCC) treated with neoadjuvant immune checkpoint blockade
- PMID: 37904980
- PMCID: PMC10614819
- DOI: 10.1101/2023.10.16.562104
Immune landscape of tertiary lymphoid structures in hepatocellular carcinoma (HCC) treated with neoadjuvant immune checkpoint blockade
Abstract
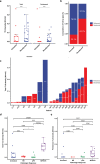
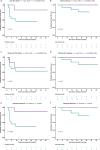
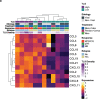
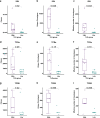
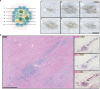
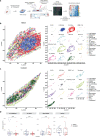
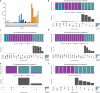
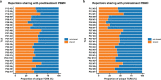
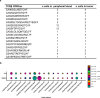
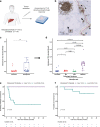
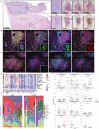
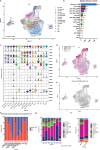
Neoadjuvant immunotherapy is thought to produce long-term remissions through induction of antitumor immune responses before removal of the primary tumor. Tertiary lymphoid structures (TLS), germinal center-like structures that can arise within tumors, may contribute to the establishment of immunological memory in this setting, but understanding of their role remains limited. Here, we investigated the contribution of TLS to antitumor immunity in hepatocellular carcinoma (HCC) treated with neoadjuvant immunotherapy. We found that neoadjuvant immunotherapy induced the formation of TLS, which were associated with superior pathologic response, improved relapse free survival, and expansion of the intratumoral T and B cell repertoire. While TLS in viable tumor displayed a highly active mature morphology, in areas of tumor regression we identified an involuted TLS morphology, which was characterized by dispersion of the B cell follicle and persistence of a T cell zone enriched for ongoing antigen presentation and T cell-mature dendritic cell interactions. Involuted TLS showed increased expression of T cell memory markers and expansion of CD8+ cytotoxic and tissue resident memory clonotypes. Collectively, these data reveal the circumstances of TLS dissolution and suggest a functional role for late-stage TLS as sites of T cell memory formation after elimination of viable tumor.
Keywords: hepatocellular carcinoma; imaging mass cytometry; immune checkpoint inhibitors; neoadjuvant immunotherapy; single cell RNA; single cell TCR; single cell multiomics; tertiary lymphoid structures.
Conflict of interest statement
Competing interests M.Y. reports grant/research support from Bristol-Myers Squibb, Incyte, Genentech (to Johns Hopkins) and honoraria from Genentech, Exelixis, Eisai, AstraZeneca, Replimune, Hepion, and equity in Adventris Pharmaceuticals. E.J.F is on the Scientific Advisory Board of Viosera/Reistance Bio, is a paid consultant for Merck and Mestag Therapeutics, and receives research funds from Abbvie. W.J.H. has received patent royalties from Rodeo/Amgen and is the recipient of grants from Sanofi, NeoTX, and CirclePharma. He has received speaking/travel honoraria from Exelixis and Standard BioTools. E.M.J. reports grant/research support from the Lustgarten Foundation, Break Through Cancer, Genentech, Bristol-Meyers Squibb; honoraria from Achilles, DragonFly, Parker Institute, Cancer Prevention and Research Institute of Texas, Surge, HDT Bio, Mestag Therapeutics, Medical Home Group; and equity in AbMeta Therapeutics and Adventris Pharmaceuticals. D.J.Z. reports grant/research support from Roche/Genentech.
Figures










References
-
- Garon EB, Hellmann MD, Rizvi NA, et al. Five-Year Overall Survival for Patients With Advanced Non‒Small-Cell Lung Cancer Treated With Pembrolizumab: Results From the Phase I KEYNOTE-001 Study. J Clin Oncol Off J Am Soc Clin Oncol. 2019;37(28):2518–2527. doi: 10.1200/JCO.19.00934 - DOI - PMC - PubMed
-
- Liu J, Blake SJ, Yong MCR, et al. Improved Efficacy of Neoadjuvant Compared to Adjuvant Immunotherapy to Eradicate Metastatic Disease. Cancer Discov. 2016;6(12):1382–1399. doi: 10.1158/2159-8290.CD-16-0577 - DOI - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
