This is a preprint.
Guanine quadruplexes mediate mitochondrial RNA polymerase pausing
- PMID: 37905021
- PMCID: PMC10614896
- DOI: 10.1101/2023.10.17.562821
Guanine quadruplexes mediate mitochondrial RNA polymerase pausing
Update in
-
Guanine quadruplexes mediate mitochondrial RNA polymerase pausing.BMC Biol. 2025 May 13;23(1):129. doi: 10.1186/s12915-025-02229-4. BMC Biol. 2025. PMID: 40361112 Free PMC article.
Abstract
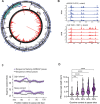
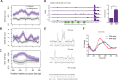
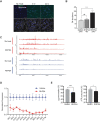
The information content within nucleic acids extends beyond the primary sequence to include secondary structures with functional roles in cells. Guanine-rich sequences form structures called guanine quadruplexes (G4) that result from non-canonical base pairing between guanine residues. These stable structures are enriched in gene promoters and have been correlated with the locations of RNA polymerase II pausing (Pol II). While promoter-proximal RNA polymerase pausing regulates gene expression, the effects of guanine quadruplexes on gene transcription have been less clear. We determined the pattern of mitochondrial RNA polymerase (mtRNAP) pausing in human fibroblasts and found that it pauses over 400 times on the mitochondrial genome. We identified quadruplexes as a mediator of mtRNAP pausing and show that stabilization of quadruplexes impeded transcription by mtRNAP. Gene products encoded by the mitochondrial genome are required for oxidative phosphorylation and the decreased transcription by mtRNAP resulted in lower expression of mitochondrial genes and significantly reduced ATP generation. Energy from mitochondria is essential for transport function in renal epithelia, and impeded mitochondrial transcription inhibits transport function in renal proximal tubule cells. These results link formation of guanine quadruplex structures to regulation of mtRNAP elongation and mitochondrial function.
Conflict of interest statement
Conflict of interests: The authors declare no competing interests.
Figures

References
-
- Anderson S., Bankier A.T., Barrell B.G., de Bruijn M.H., Coulson A.R., Drouin J., Eperon I.C., Nierlich D.P., Roe B.A., Sanger F. et al. (1981) Sequence and organization of the human mitochondrial genome. Nature, 290, 457–465. - PubMed
-
- Ojala D., Montoya J. and Attardi G. (1981) tRNA punctuation model of RNA processing in human mitochondria. Nature, 290, 470–474. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
