This is a preprint.
hkb is required for DIP-α expression and target recognition in the Drosophila neuromuscular circuit
- PMID: 37905128
- PMCID: PMC10614772
- DOI: 10.1101/2023.10.15.562341
hkb is required for DIP-α expression and target recognition in the Drosophila neuromuscular circuit
Update in
-
hkb is required for DIP-α expression and target recognition in the Drosophila neuromuscular circuit.Commun Biol. 2024 Apr 27;7(1):507. doi: 10.1038/s42003-024-06184-8. Commun Biol. 2024. PMID: 38678127 Free PMC article.
Abstract
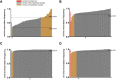
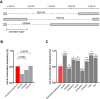
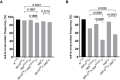
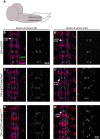
Our nervous system contains billions of neurons that form precise connections with each other through interactions between cell surface proteins (CSPs). In Drosophila, the Dpr and DIP immunoglobulin protein subfamilies form homophilic or heterophilic interactions to instruct synaptic connectivity, synaptic growth and cell survival. However, the upstream regulation and downstream signaling mechanisms of Dprs and DIPs are not clear. In the Drosophila larval neuromuscular system, DIP-α is expressed in the dorsal and ventral type-Is motor neurons (MNs). We conducted an F1 dominant modifier genetic screen to identify regulators of Dprs and DIPs. We found that the transcription factor, huckebein (hkb), genetically interacts with DIP-α and is important for target recognition specifically in the dorsal Is MN, but not the ventral Is MN. Loss of hkb led to complete removal of DIP-α expression. We then confirmed that this specificity is through the dorsal Is MN specific transcription factor, even-skipped (eve), which acts downstream of hkb. Genetic interaction between hkb and eve revealed that they act in the same pathway to regulate dorsal Is MN connectivity. Our study provides insight into the transcriptional regulation of DIP-α and suggests that distinct regulatory mechanisms exist for the same CSP in different neurons.
Figures

References
-
- Bossing T., Technau G. M. and Doe C. Q. (1996). huckebein is required for glial development and axon pathfinding in the neuroblast 1–1 and neuroblast 2–2 lineages in the Drosophila central nervous system. Mech Develop 55, 53–64. - PubMed
-
- Broadus J., Skeath J. B., Spana E. P., Bossing T., Technau G. and Doe C. Q. (1995). New neuroblast markers and the origin of the aCC/pCC neurons in the Drosophila central nervous system. Mech Develop 53, 393–402. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
