Loss of the methylarginine reader function of SND1 confers resistance to hepatocellular carcinoma
- PMID: 37905668
- PMCID: PMC10860161
- DOI: 10.1042/BCJ20230384
Loss of the methylarginine reader function of SND1 confers resistance to hepatocellular carcinoma
Abstract
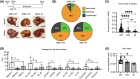
Staphylococcal nuclease Tudor domain containing 1 (SND1) protein is an oncogene that 'reads' methylarginine marks through its Tudor domain. Specifically, it recognizes methylation marks deposited by protein arginine methyltransferase 5 (PRMT5), which is also known to promote tumorigenesis. Although SND1 can drive hepatocellular carcinoma (HCC), it is unclear whether the SND1 Tudor domain is needed to promote HCC. We sought to identify the biological role of the SND1 Tudor domain in normal and tumorigenic settings by developing two genetically engineered SND1 mouse models, an Snd1 knockout (Snd1 KO) and an Snd1 Tudor domain-mutated (Snd1 KI) mouse, whose mutant SND1 can no longer recognize PRMT5-catalyzed methylarginine marks. Quantitative PCR analysis of normal, KO, and KI liver samples revealed a role for the SND1 Tudor domain in regulating the expression of genes encoding major acute phase proteins, which could provide mechanistic insight into SND1 function in a tumor setting. Prior studies indicated that ectopic overexpression of SND1 in the mouse liver dramatically accelerates the development of diethylnitrosamine (DEN)-induced HCC. Thus, we tested the combined effects of DEN and SND1 loss or mutation on the development of HCC. We found that both Snd1 KO and Snd1 KI mice were partially protected against malignant tumor development following exposure to DEN. These results support the development of small molecule inhibitors that target the SND1 Tudor domain or the use of upstream PRMT5 inhibitors, as novel treatments for HCC.
Keywords: HCC; SND1; arginine methylation; liver cancer; tudor domain.
© 2023 The Author(s).
Conflict of interest statement
Mark T. Bedford is the co-founder of EpiCypher.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

