Impact of AlphaFold on structure prediction of protein complexes: The CASP15-CAPRI experiment
- PMID: 37905971
- PMCID: PMC10841881
- DOI: 10.1002/prot.26609
Impact of AlphaFold on structure prediction of protein complexes: The CASP15-CAPRI experiment
Abstract
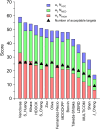
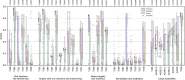
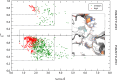
We present the results for CAPRI Round 54, the 5th joint CASP-CAPRI protein assembly prediction challenge. The Round offered 37 targets, including 14 homodimers, 3 homo-trimers, 13 heterodimers including 3 antibody-antigen complexes, and 7 large assemblies. On average ~70 CASP and CAPRI predictor groups, including more than 20 automatics servers, submitted models for each target. A total of 21 941 models submitted by these groups and by 15 CAPRI scorer groups were evaluated using the CAPRI model quality measures and the DockQ score consolidating these measures. The prediction performance was quantified by a weighted score based on the number of models of acceptable quality or higher submitted by each group among their five best models. Results show substantial progress achieved across a significant fraction of the 60+ participating groups. High-quality models were produced for about 40% of the targets compared to 8% two years earlier. This remarkable improvement is due to the wide use of the AlphaFold2 and AlphaFold2-Multimer software and the confidence metrics they provide. Notably, expanded sampling of candidate solutions by manipulating these deep learning inference engines, enriching multiple sequence alignments, or integration of advanced modeling tools, enabled top performing groups to exceed the performance of a standard AlphaFold2-Multimer version used as a yard stick. This notwithstanding, performance remained poor for complexes with antibodies and nanobodies, where evolutionary relationships between the binding partners are lacking, and for complexes featuring conformational flexibility, clearly indicating that the prediction of protein complexes remains a challenging problem.
Keywords: AlphaFold; CAPRI; CASP; blind prediction; deep learning; protein assemblies; protein complexes; protein-protein interaction.
© 2023 The Authors. Proteins: Structure, Function, and Bioinformatics published by Wiley Periodicals LLC.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




References
-
- Alberts B. The cell as a collection of protein machines: preparing the next generation of molecular biologists. Cell. 1998;92(3):291‐294. - PubMed
-
- Berman H, Henrick K, Nakamura H. Announcing the worldwide Protein Data Bank. Nat Struct Biol. 2003;10(12):980. - PubMed
-
- Bai XC, McMullan G, Scheres SH. How cryo‐EM is revolutionizing structural biology. Trends Biochem Sci. 2015;40(1):49‐57. - PubMed
Publication types
MeSH terms
Grants and funding
- R35 GM136409/GM/NIGMS NIH HHS/United States
- R01 GM140098/GM/NIGMS NIH HHS/United States
- R35 GM141881/GM/NIGMS NIH HHS/United States
- R35 GM118078/GM/NIGMS NIH HHS/United States
- R35 GM124952/GM/NIGMS NIH HHS/United States
- FC0001003/CRUK_/Cancer Research UK/United Kingdom
- FC001003/WT_/Wellcome Trust/United Kingdom
- R35 GM144083/GM/NIGMS NIH HHS/United States
- R01 GM146340/GM/NIGMS NIH HHS/United States
- R01 GM123055/GM/NIGMS NIH HHS/United States
- R43 GM144992/GM/NIGMS NIH HHS/United States
- T32 GM132024/GM/NIGMS NIH HHS/United States
- R01 GM133840/GM/NIGMS NIH HHS/United States
- R01 GM093123/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources

