Diabetes-associated breast cancer is molecularly distinct and shows a DNA damage repair deficiency
- PMID: 37906280
- PMCID: PMC10795835
- DOI: 10.1172/jci.insight.170105
Diabetes-associated breast cancer is molecularly distinct and shows a DNA damage repair deficiency
Abstract
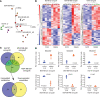
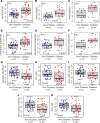
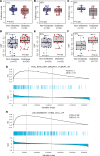
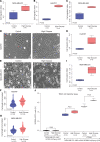
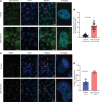
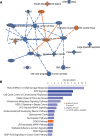
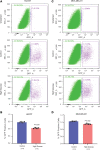
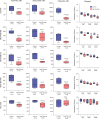
Diabetes commonly affects patients with cancer. We investigated the influence of diabetes on breast cancer biology using a 3-pronged approach that included analysis of orthotopic human tumor xenografts, patient tumors, and breast cancer cells exposed to diabetes/hyperglycemia-like conditions. We aimed to identify shared phenotypes and molecular signatures by investigating the metabolome, transcriptome, and tumor mutational burden. Diabetes and hyperglycemia did not enhance cell proliferation but induced mesenchymal and stem cell-like phenotypes linked to increased mobility and odds of metastasis. They also promoted oxyradical formation and both a transcriptome and mutational signatures of DNA repair deficiency. Moreover, food- and microbiome-derived metabolites tended to accumulate in breast tumors in the presence of diabetes, potentially affecting tumor biology. Breast cancer cells cultured under hyperglycemia-like conditions acquired increased DNA damage and sensitivity to DNA repair inhibitors. Based on these observations, we conclude that diabetes-associated breast tumors may show an increased drug response to DNA damage repair inhibitors.
Keywords: Breast cancer; Diabetes; Metabolism; Oncology.
Conflict of interest statement
Figures




References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

