A genome-scale metabolic model of parasitic whipworm
- PMID: 37907472
- PMCID: PMC10618284
- DOI: 10.1038/s41467-023-42552-4
A genome-scale metabolic model of parasitic whipworm
Abstract
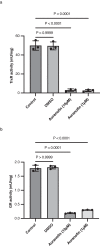
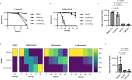
Genome-scale metabolic models are widely used to enhance our understanding of metabolic features of organisms, host-pathogen interactions and to identify therapeutics for diseases. Here we present iTMU798, the genome-scale metabolic model of the mouse whipworm Trichuris muris. The model demonstrates the metabolic features of T. muris and allows the prediction of metabolic steps essential for its survival. Specifically, that Thioredoxin Reductase (TrxR) enzyme is essential, a prediction we validate in vitro with the drug auranofin. Furthermore, our observation that the T. muris genome lacks gsr-1 encoding Glutathione Reductase (GR) but has GR activity that can be inhibited by auranofin indicates a mechanism for the reduction of glutathione by the TrxR enzyme in T. muris. In addition, iTMU798 predicts seven essential amino acids that cannot be synthesised by T. muris, a prediction we validate for the amino acid tryptophan. Overall, iTMU798 is as a powerful tool to study not only the T. muris metabolism but also other Trichuris spp. in understanding host parasite interactions and the rationale design of new intervention strategies.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- WHO. Soil-transmitted Helminth Infections. https://www.who.int/news-room/fact-sheets/detail/soil-transmitted-helmin... (2022).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

