Loss of Grem1-lineage chondrogenic progenitor cells causes osteoarthritis
- PMID: 37907525
- PMCID: PMC10618187
- DOI: 10.1038/s41467-023-42199-1
Loss of Grem1-lineage chondrogenic progenitor cells causes osteoarthritis
Abstract
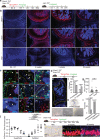
Osteoarthritis (OA) is characterised by an irreversible degeneration of articular cartilage. Here we show that the BMP-antagonist Gremlin 1 (Grem1) marks a bipotent chondrogenic and osteogenic progenitor cell population within the articular surface. Notably, these progenitors are depleted by injury-induced OA and increasing age. OA is also caused by ablation of Grem1 cells in mice. Transcriptomic and functional analysis in mice found that articular surface Grem1-lineage cells are dependent on Foxo1 and ablation of Foxo1 in Grem1-lineage cells caused OA. FGFR3 signalling was confirmed as a promising therapeutic pathway by administration of pathway activator, FGF18, resulting in Grem1-lineage chondrocyte progenitor cell proliferation, increased cartilage thickness and reduced OA. These findings suggest that OA, in part, is caused by mechanical, developmental or age-related attrition of Grem1 expressing articular cartilage progenitor cells. These cells, and the FGFR3 signalling pathway that sustains them, may be effective future targets for biological management of OA.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

