Early-life origin of prostate cancer through deregulation of miR-206 networks in maternally malnourished offspring rats
- PMID: 37907720
- PMCID: PMC10618455
- DOI: 10.1038/s41598-023-46068-1
Early-life origin of prostate cancer through deregulation of miR-206 networks in maternally malnourished offspring rats
Abstract
The Developmental Origins of Health and Disease (DOHaD) concept has provided the framework to assess how early life experiences can shape health and disease throughout the life course. While maternal malnutrition has been proposed as a risk factor for the developmental programming of prostate cancer (PCa), the molecular mechanisms remain poorly understood. Using RNA-seq data, we demonstrated deregulation of miR-206-Plasminogen (PLG) network in the ventral prostate (VP) of young maternally malnourished offspring. RT-qPCR confirmed the deregulation of the miR-206-PLG network in the VP of young and old offspring rats. Considering the key role of estrogenic signaling pathways in prostate carcinogenesis, in vitro miRNA mimic studies also revealed a negative correlation between miR-206 and estrogen receptor α (ESR1) expression in PNT2 cells. Together, we demonstrate that early life estrogenization associated with the deregulation of miR-206 networks can contribute to the developmental origins of PCa in maternally malnourished offspring. Understanding the molecular mechanisms by which early life malnutrition affects offspring health can encourage the adoption of a governmental policy for the prevention of non-communicable chronic diseases related to the DOHaD concept.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
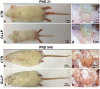
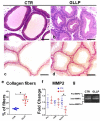
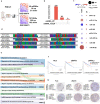

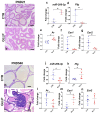

References
-
- WHO, W. H. O. World Health Organization, Health Topics: Nutrition. (2021). Available at: https://www.who.int/health-topics/nutrition. (Accessed: 31st August 2021).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

