Microbial hitchhikers harbouring antimicrobial-resistance genes in the riverine plastisphere
- PMID: 37908022
- PMCID: PMC10619285
- DOI: 10.1186/s40168-023-01662-3
Microbial hitchhikers harbouring antimicrobial-resistance genes in the riverine plastisphere
Abstract
Background: The widespread nature of plastic pollution has given rise to wide scientific and social concern regarding the capacity of these materials to serve as vectors for pathogenic bacteria and reservoirs for Antimicrobial Resistance Genes (ARG). In- and ex-situ incubations were used to characterise the riverine plastisphere taxonomically and functionally in order to determine whether antibiotics within the water influenced the ARG profiles in these microbiomes and how these compared to those on natural surfaces such as wood and their planktonic counterparts.
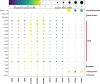
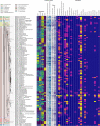
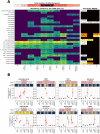
Results: We show that plastics support a taxonomically distinct microbiome containing potential pathogens and ARGs. While the plastisphere was similar to those biofilms that grew on wood, they were distinct from the surrounding water microbiome. Hence, whilst potential opportunistic pathogens (i.e. Pseudomonas aeruginosa, Acinetobacter and Aeromonas) and ARG subtypes (i.e. those that confer resistance to macrolides/lincosamides, rifamycin, sulfonamides, disinfecting agents and glycopeptides) were predominant in all surface-related microbiomes, especially on weathered plastics, a completely different set of potential pathogens (i.e. Escherichia, Salmonella, Klebsiella and Streptococcus) and ARGs (i.e. aminoglycosides, tetracycline, aminocoumarin, fluoroquinolones, nitroimidazole, oxazolidinone and fosfomycin) dominated in the planktonic compartment. Our genome-centric analysis allowed the assembly of 215 Metagenome Assembled Genomes (MAGs), linking ARGs and other virulence-related genes to their host. Interestingly, a MAG belonging to Escherichia -that clearly predominated in water- harboured more ARGs and virulence factors than any other MAG, emphasising the potential virulent nature of these pathogenic-related groups. Finally, ex-situ incubations using environmentally-relevant concentrations of antibiotics increased the prevalence of their corresponding ARGs, but different riverine compartments -including plastispheres- were affected differently by each antibiotic.
Conclusions: Our results provide insights into the capacity of the riverine plastisphere to harbour a distinct set of potentially pathogenic bacteria and function as a reservoir of ARGs. The environmental impact that plastics pose if they act as a reservoir for either pathogenic bacteria or ARGs is aggravated by the persistence of plastics in the environment due to their recalcitrance and buoyancy. Nevertheless, the high similarities with microbiomes growing on natural co-occurring materials and even more worrisome microbiome observed in the surrounding water highlights the urgent need to integrate the analysis of all environmental compartments when assessing risks and exposure to pathogens and ARGs in anthropogenically-impacted ecosystems. Video Abstract.
© 2023. BioMed Central Ltd., part of Springer Nature.
Conflict of interest statement
E.M.H.W. and J.A.C-O are currently serving as the Editor-in-Chief and Associate Editor, respectively, for the journal Microbiome. However, it is important to note that they were not involved in the Handling Editor or Reviewer selection processes, nor any other stage of the evaluation and decision process.
Figures


References
-
- Alcock BP, Raphenya AR, Lau TTY, Tsang KK, Bouchard M, Edalatmand A, Huynh W, Nguyen ALV, Cheng AA, Liu S, Min SY, Miroshnichenko A, Tran HK, Werfalli RE, Nasir JA, Oloni M, Speicher DJ, Florescu A, Singh B, Faltyn M, Hernandez-Koutoucheva A, Sharma AN, Bordeleau E, Pawlowski AC, Zubyk HL, Dooley D, Griffiths E, Maguire F, Winsor GL, Beiko RG, Brinkman FSL, Hsiao WWL, Domselaar GV, McArthur AG. CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020;48:D517–D525. doi: 10.1093/nar/gkz935. - DOI - PMC - PubMed
-
- Allaire J, Ushey K, Tang Y, Eddelbuettel D. reticulate: R Interface to Python. 2017.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

