Structural properties of the HNF-1A transactivation domain
- PMID: 37908230
- PMCID: PMC10613711
- DOI: 10.3389/fmolb.2023.1249939
Structural properties of the HNF-1A transactivation domain
Abstract
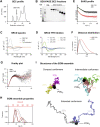
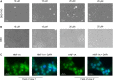
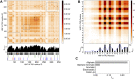
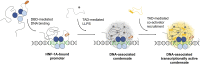
Hepatocyte nuclear factor 1α (HNF-1A) is a transcription factor with important gene regulatory roles in pancreatic β-cells. HNF1A gene variants are associated with a monogenic form of diabetes (HNF1A-MODY) or an increased risk for type 2 diabetes. While several pancreatic target genes of HNF-1A have been described, a lack of knowledge regarding the structure-function relationships in HNF-1A prohibits a detailed understanding of HNF-1A-mediated gene transcription, which is important for precision medicine and improved patient care. Therefore, we aimed to characterize the understudied transactivation domain (TAD) of HNF-1A in vitro. We present a bioinformatic approach to dissect the TAD sequence, analyzing protein structure, sequence composition, sequence conservation, and the existence of protein interaction motifs. Moreover, we developed the first protocol for the recombinant expression and purification of the HNF-1A TAD. Small-angle X-ray scattering and synchrotron radiation circular dichroism suggested a disordered conformation for the TAD. Furthermore, we present functional data on HNF-1A undergoing liquid-liquid phase separation, which is in line with in silico predictions and may be of biological relevance for gene transcriptional processes in pancreatic β-cells.
Keywords: HNF-1A; MODY; diabetes; intrinsically disordered protein; liquid-liquid phase separation; short linear motif; transcription factor; β-cell.
Copyright © 2023 Kind, Driver, Raasakka, Onck, Njølstad, Arnesen and Kursula.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
LinkOut - more resources
Full Text Sources

