Metabolism of pancreatic neuroendocrine tumors: what can omics tell us?
- PMID: 37908747
- PMCID: PMC10613989
- DOI: 10.3389/fendo.2023.1248575
Metabolism of pancreatic neuroendocrine tumors: what can omics tell us?
Abstract
Introduction: Reprogramming of cellular metabolism is now a hallmark of tumorigenesis. In recent years, research on pancreatic neuroendocrine tumors (pNETs) has focused on genetic and epigenetic modifications and related signaling pathways, but few studies have been devoted to characterizing the metabolic profile of these tumors. In this review, we thoroughly investigate the metabolic pathways in pNETs by analyzing the transcriptomic and metabolomic data available in the literature.
Methodology: We retrieved and downloaded gene expression profiles from all publicly available gene set enrichments (GSE43797, GSE73338, and GSE117851) to compare the differences in expressed genes based on both the stage and MEN1 mutational status. In addition, we conducted a systematic review of metabolomic data in NETs.
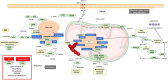
Results: By combining transcriptomic and metabolomic approaches, we have identified a distinctive metabolism in pNETs compared with controls without pNETs. Our analysis showed dysregulations in the one-carbon, glutathione, and polyamine metabolisms, fatty acid biosynthesis, and branched-chain amino acid catabolism, which supply the tricarboxylic acid cycle. These targets are implicated in pNET cell proliferation and metastasis and could also have a prognostic impact. When analyzing the profiles of patients with or without metastasis, or with or without MEN1 mutation, we observed only a few differences due to the scarcity of published clinical data in the existing research. Consequently, further studies are now necessary to validate our data and investigate these potential targets as biomarkers or therapeutic solutions, with a specific focus on pNETs.
Keywords: MEN1; integrative biology; metabolism; metabolomic; metastasis; pancreatic neuroendocrine tumors; transcriptomic.
Copyright © 2023 Jannin, Dessein, Do Cao, Vantyghem, Chevalier, Van Seuningen, Jonckheere and Coppin.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
-
- Hanahan D. Hallmarks of cancer: new dimensions. Cancer Discov (2022) 12(1):31–46. doi: 10.1158/2159-8290.CD-21-1059 - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

