Chemical Proteomic Discovery of Isotype-Selective Covalent Inhibitors of the RNA Methyltransferase NSUN2
- PMID: 37909922
- PMCID: PMC10999112
- DOI: 10.1002/anie.202311924
Chemical Proteomic Discovery of Isotype-Selective Covalent Inhibitors of the RNA Methyltransferase NSUN2
Abstract
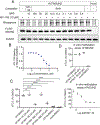
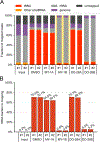
5-Methylcytosine (m5 C) is an RNA modification prevalent on tRNAs, where it can protect tRNAs from endonucleolytic cleavage to maintain protein synthesis. The NSUN family (NSUN1-7 in humans) of RNA methyltransferases are capable of installing the methyl group onto the C5 position of cytosines in RNA. NSUNs are implicated in a wide range of (patho)physiological processes, but selective and cell-active inhibitors of these enzymes are lacking. Here, we use cysteine-directed activity-based protein profiling (ABPP) to discover azetidine acrylamides that act as stereoselective covalent inhibitors of human NSUN2. Despite targeting a conserved catalytic cysteine in the NSUN family, the NSUN2 inhibitors show negligible cross-reactivity with other human NSUNs and exhibit good proteome-wide selectivity. We verify that the azetidine acrylamides inhibit the catalytic activity of recombinant NSUN2, but not NSUN6, and demonstrate that these compounds stereoselectively disrupt NSUN2-tRNA interactions in cancer cells, leading to a global reduction in tRNA m5 C content. Our findings thus highlight the potential to create isotype-selective and cell-active inhibitors of NSUN2 with covalent chemistry targeting a conserved catalytic cysteine.
Keywords: 5-Methylcytosine; Activity-Based Protein Profiling; Covalent Inhibitor; NSUN2; RNA Methylation.
© 2023 The Authors. Angewandte Chemie International Edition published by Wiley-VCH GmbH.
Conflict of interest statement
Competing interests
Dr. Cravatt is a founder and advisor to Vividion Therapeutics. C.H. is a scientific founder, a member of the scientific advisory board and equity holder of Aferna Green, Inc. and AccuaDX Inc., and a scientific co-founder and equity holder of Accent Therapeutics, Inc. Drs. Cravatt, Melillo, and Schreiber are founders and/or scientific advisors to Magnet Therapeutics.
Figures




References
-
- Thüring K, Schmid K, Keller P, Helm M, Methods 2016, 107, 48–56. - PubMed
-
- a) Motorin Y, Lyko F, Helm M, Nucleic Acids Res. 2010, 38, 1415–1430; - PMC - PubMed
- b)Guo G, Pan K, Fang S, Ye L, Tong X, Wang Z, Xue X, Zhang H, Mol. Ther. Nucleic. Acids 2021, 26, 575–593; - PMC - PubMed
- c)Squires JE, Preiss T, Epigenomics 2010, 2, 709–715; - PubMed
- d)Trixl L, Lusser A, Wiley Interdiscip. Rev. RNA 2019, 10, e1510. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

