Hepatocyte FBXW7-dependent activity of nutrient-sensing nuclear receptors controls systemic energy homeostasis and NASH progression in male mice
- PMID: 37914694
- PMCID: PMC10620240
- DOI: 10.1038/s41467-023-42785-3
Hepatocyte FBXW7-dependent activity of nutrient-sensing nuclear receptors controls systemic energy homeostasis and NASH progression in male mice
Abstract
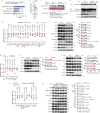
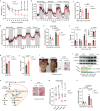
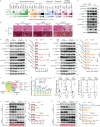
Nonalcoholic steatohepatitis (NASH) is epidemiologically associated with obesity and diabetes and can lead to liver cirrhosis and hepatocellular carcinoma if left untreated. The intricate signaling pathways that orchestrate hepatocyte energy metabolism and cellular stress, intrahepatic cell crosstalk, as well as interplay between peripheral tissues remain elusive and are crucial for the development of anti-NASH therapies. Herein, we reveal E3 ligase FBXW7 as a key factor regulating hepatic catabolism, stress responses, systemic energy homeostasis, and NASH pathogenesis with attenuated FBXW7 expression as a feature of advanced NASH. Multiomics and pharmacological intervention showed that FBXW7 loss-of-function in hepatocytes disrupts a metabolic transcriptional axis conjointly controlled by the nutrient-sensing nuclear receptors ERRα and PPARα, resulting in suppression of fatty acid oxidation, elevated ER stress, apoptosis, immune infiltration, fibrogenesis, and ultimately NASH progression in male mice. These results provide the foundation for developing alternative strategies co-targeting ERRα and PPARα for the treatment of NASH.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

