Mouse genome rewriting and tailoring of three important disease loci
- PMID: 37914927
- PMCID: PMC10632133
- DOI: 10.1038/s41586-023-06675-4
Mouse genome rewriting and tailoring of three important disease loci
Abstract
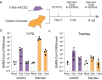
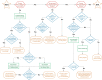
Genetically engineered mouse models (GEMMs) help us to understand human pathologies and develop new therapies, yet faithfully recapitulating human diseases in mice is challenging. Advances in genomics have highlighted the importance of non-coding regulatory genome sequences, which control spatiotemporal gene expression patterns and splicing in many human diseases1,2. Including regulatory extensive genomic regions, which requires large-scale genome engineering, should enhance the quality of disease modelling. Existing methods set limits on the size and efficiency of DNA delivery, hampering the routine creation of highly informative models that we call genomically rewritten and tailored GEMMs (GREAT-GEMMs). Here we describe 'mammalian switching antibiotic resistance markers progressively for integration' (mSwAP-In), a method for efficient genome rewriting in mouse embryonic stem cells. We demonstrate the use of mSwAP-In for iterative genome rewriting of up to 115 kb of a tailored Trp53 locus, as well as for humanization of mice using 116 kb and 180 kb human ACE2 loci. The ACE2 model recapitulated human ACE2 expression patterns and splicing, and notably, presented milder symptoms when challenged with SARS-CoV-2 compared with the existing K18-hACE2 model, thus representing a more human-like model of infection. Finally, we demonstrated serial genome writing by humanizing mouse Tmprss2 biallelically in the ACE2 GREAT-GEMM, highlighting the versatility of mSwAP-In in genome writing.
© 2023. The Author(s).
Conflict of interest statement
J.D.B. is a founder and director of CDI Labs, a founder of and consultant to Neochromosome, a founder, scientific advisory board member of and consultant to ReOpen Diagnostics, and serves or served on the scientific advisory board of the following: Logomix, Modern Meadow, Rome Therapeutics, Sample6, Sangamo, Tessera Therapeutics and the Wyss Institute. The mSwAP-In method described here is the subject of a pending patent application. The other authors declare no competing interests.
Figures
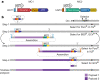


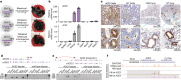


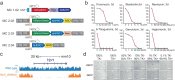





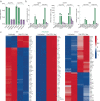


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

