Bayesian traction force estimation using cell boundary-dependent force priors
- PMID: 37915171
- PMCID: PMC10719052
- DOI: 10.1016/j.bpj.2023.10.032
Bayesian traction force estimation using cell boundary-dependent force priors
Abstract
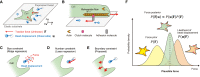
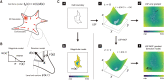
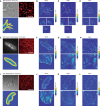
Understanding the principles of cell migration necessitates measurements of the forces generated by cells. In traction force microscopy (TFM), fluorescent beads are placed on a substrate's surface and the substrate strain caused by the cell traction force is observed as displacement of the beads. Mathematical analysis can estimate traction force from bead displacement. However, most algorithms estimate substrate stresses independently of cell boundary, which results in poor estimation accuracy in low-density bead environments. To achieve accurate force estimation at low density, we proposed a Bayesian traction force estimation (BTFE) algorithm that incorporates cell-boundary-dependent force as a prior. We evaluated the performance of the proposed algorithm using synthetic data generated with mathematical models of cells and TFM substrates. BTFE outperformed other methods, especially in low-density bead conditions. In addition, the BTFE algorithm provided a reasonable force estimation using TFM images from the experiment.
Copyright © 2023 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



References
-
- Calandra T., Bucala R. Macrophage migration inhibitory factor (MIF): a glucocorticoid counter-regulator within the immune system. Crit. Rev. Immunol. 1997;17:77–88. - PubMed
-
- Luster A.D., Alon R., von Andrian U.H. Immune cell migration in inflammation: present and future therapeutic targets. Nat. Immunol. 2005;6:1182–1190. - PubMed
-
- Tessier-Lavigne M., Goodman C.S. The molecular biology of axon guidance. Science. 1996;274:1123–1133. - PubMed
-
- Dickson B.J. Molecular mechanisms of axon guidance. Science. 2002;298:1959–1964. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

