Recapitulation of Perturbed Striatal Gene Expression Dynamics of Donors' Brains With Ventral Forebrain Organoids Derived From the Same Individuals With Schizophrenia
- PMID: 37915216
- PMCID: PMC11209846
- DOI: 10.1176/appi.ajp.20220723
Recapitulation of Perturbed Striatal Gene Expression Dynamics of Donors' Brains With Ventral Forebrain Organoids Derived From the Same Individuals With Schizophrenia
Abstract
Objective: Schizophrenia is a brain disorder that originates during neurodevelopment and has complex genetic and environmental etiologies. Despite decades of clinical evidence of altered striatal function in affected patients, studies examining its cellular and molecular mechanisms in humans are limited. To explore neurodevelopmental alterations in the striatum associated with schizophrenia, the authors established a method for the differentiation of induced pluripotent stem cells (iPSCs) into ventral forebrain organoids (VFOs).
Methods: VFOs were generated from postmortem dural fibroblast-derived iPSCs of four individuals with schizophrenia and four neurotypical control individuals for whom postmortem caudate genotypes and transcriptomic data were profiled in the BrainSeq neurogenomics consortium. Individuals were selected such that the two groups had nonoverlapping schizophrenia polygenic risk scores (PRSs).
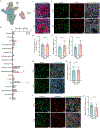
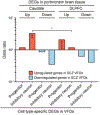
Results: Single-cell RNA sequencing analyses of VFOs revealed differences in developmental trajectory between schizophrenia and control individuals in which inhibitory neuronal cells from the patients exhibited accelerated maturation. Furthermore, upregulated genes in inhibitory neurons in schizophrenia VFOs showed a significant overlap with upregulated genes in postmortem caudate tissue of individuals with schizophrenia compared with control individuals, including the donors of the iPSC cohort.
Conclusions: The findings suggest that striatal neurons derived from high-PRS individuals with schizophrenia carry abnormalities that originated during early brain development and that the VFO model can recapitulate disease-relevant cell type-specific neurodevelopmental phenotypes in a dish.
Keywords: Genetics/Genomics; Schizophrenia Spectrum and Other Psychotic Disorders.
Conflict of interest statement
Conflict of Interest
DRW serves on the advisory boards of Sage Therapeutics and Pasithea Therapeutics. The other authors declare no conflict of interest.
Figures



References
-
- Jablensky A, Kirkbride JB & Jones PB. (2011). Schizophrenia: The epidemiological horizon. In: Weinberger DR & Harrison PJ (eds). Schizophrenia. Wiley-Blackwell: New Jersey, pp 185–225.
-
- Kahn RS, Sommer IE, Murray RM, et al. Schizophrenia. Nat Rev Dis Primers. 2015; 1: 15067. - PubMed
-
- Birnbaum R & Weinberger DR. Genetic insights into the neurodevelopmental origins of schizophrenia. Nat. Rev. Neurosci 2017; 18, 727–740. - PubMed
-
- Millan MJ, Andrieux A, Bartzokis G, et al. Altering the course of schizophrenia: progress and perspectives. Nat Rev Drug Discov. 2016; 15: 485–515. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

