SEGS-1 a cassava genomic sequence increases the severity of African cassava mosaic virus infection in Arabidopsis thaliana
- PMID: 37915512
- PMCID: PMC10616593
- DOI: 10.3389/fpls.2023.1250105
SEGS-1 a cassava genomic sequence increases the severity of African cassava mosaic virus infection in Arabidopsis thaliana
Abstract
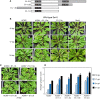
Cassava is a major crop in Sub-Saharan Africa, where it is grown primarily by smallholder farmers. Cassava production is constrained by Cassava mosaic disease (CMD), which is caused by a complex of cassava mosaic begomoviruses (CMBs). A previous study showed that SEGS-1 (sequences enhancing geminivirus symptoms), which occurs in the cassava genome and as episomes during viral infection, enhances CMD symptoms and breaks resistance in cassava. We report here that SEGS-1 also increases viral disease severity in Arabidopsis thaliana plants that are co-inoculated with African cassava mosaic virus (ACMV) and SEGS-1 sequences. Viral disease was also enhanced in Arabidopsis plants carrying a SEGS-1 transgene when inoculated with ACMV alone. Unlike cassava, no SEGS-1 episomal DNA was detected in the transgenic Arabidopsis plants during ACMV infection. Studies using Nicotiana tabacum suspension cells showed that co-transfection of SEGS-1 sequences with an ACMV replicon increases viral DNA accumulation in the absence of viral movement. Together, these results demonstrated that SEGS-1 can function in a heterologous host to increase disease severity. Moreover, SEGS-1 is active in a host genomic context, indicating that SEGS-1 episomes are not required for disease enhancement.
Keywords: ACMV; Arabidopsis thaliana; SEGS-1; begomovirus; cassava.
Copyright © 2023 Rajabu, Dallas, Chiunga, De León, Ateka, Tairo, Ndunguru, Ascencio-Ibanez and Hanley-Bowdoin.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
SEGS-1 episomes generated during cassava mosaic disease enhance disease severity.Front Plant Sci. 2025 Jan 10;15:1469045. doi: 10.3389/fpls.2024.1469045. eCollection 2024. Front Plant Sci. 2025. PMID: 39911651 Free PMC article.
-
A New Type of Satellite Associated with Cassava Mosaic Begomoviruses.J Virol. 2021 Oct 13;95(21):e0043221. doi: 10.1128/JVI.00432-21. Epub 2021 Aug 18. J Virol. 2021. PMID: 34406866 Free PMC article.
-
Two Novel DNAs That Enhance Symptoms and Overcome CMD2 Resistance to Cassava Mosaic Disease.J Virol. 2016 Mar 28;90(8):4160-4173. doi: 10.1128/JVI.02834-15. Print 2016 Apr. J Virol. 2016. PMID: 26865712 Free PMC article.
-
Cassava Mosaic and Brown Streak Diseases: Current Perspectives and Beyond.Annu Rev Virol. 2017 Sep 29;4(1):429-452. doi: 10.1146/annurev-virology-101416-041913. Epub 2017 Jun 23. Annu Rev Virol. 2017. PMID: 28645239 Review.
-
The Search for Resistance to Cassava Mosaic Geminiviruses: How Much We Have Accomplished, and What Lies Ahead.Front Plant Sci. 2017 Mar 24;8:408. doi: 10.3389/fpls.2017.00408. eCollection 2017. Front Plant Sci. 2017. PMID: 28392798 Free PMC article. Review.
Cited by
-
SEGS-1 episomes generated during cassava mosaic disease enhance disease severity.Front Plant Sci. 2025 Jan 10;15:1469045. doi: 10.3389/fpls.2024.1469045. eCollection 2024. Front Plant Sci. 2025. PMID: 39911651 Free PMC article.
-
DNA Satellites Impact Begomovirus Diseases in a Virus-Specific Manner.Int J Mol Sci. 2025 Jun 17;26(12):5814. doi: 10.3390/ijms26125814. Int J Mol Sci. 2025. PMID: 40565275 Free PMC article. Review.
References
-
- Abbas Q., Amin I., Mansoor S., Shafiq M., Wassenegger M., Briddon R. W. (2019). The Rep proteins encoded by alphasatellites restore expression of a transcriptionally silenced green fluorescent protein transgene in Nicotiana benthamiana. Virusdisease 30, 101–105. doi: 10.1007/s13337-017-0413-5 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

