Single-cell profiling of glial cells from the mouse amygdala under opioid dependent and withdrawal states
- PMID: 37915593
- PMCID: PMC10616319
- DOI: 10.1016/j.isci.2023.108166
Single-cell profiling of glial cells from the mouse amygdala under opioid dependent and withdrawal states
Abstract
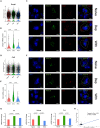
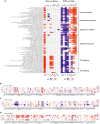
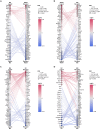
The cycle of substance use disorder (SUD) leading to dependence is a complex process involving multiple neurocircuitries and brain regions. The amygdala is the core brain region that is involved in processing withdrawal and anxiety and depressive-like behaviors. However, the transcriptional changes in each cell type within the amygdala during SUD remains unknown. Here, we performed single-cell RNA sequencing and classified all cell types in the mouse amygdala. We particularly focused on gene expression changes in glial cells under dependent state and compared to either naive or withdrawal state. Our data revealed distinct changes in key biological processes, such as gene expression, immune response, inflammation, synaptic transmission, and mitochondrial respiration. Significant differences were unraveled in the transcriptional profiles between dependence and withdrawal states. This report is the first single-cell RNA sequencing dataset from the amygdala under opioid dependence and withdrawal conditions, providing unique insights in understanding brain alterations during SUD.
Keywords: Neuroscience; Transcriptomics.
© 2023 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

