Unbiased assessment of genome integrity and purging of adverse outcomes at the target locus upon editing of CD4+ T-cells for the treatment of Hyper IgM1
- PMID: 37916874
- PMCID: PMC10690452
- DOI: 10.15252/embj.2023114188
Unbiased assessment of genome integrity and purging of adverse outcomes at the target locus upon editing of CD4+ T-cells for the treatment of Hyper IgM1
Abstract
Hyper IgM1 is an X-linked combined immunodeficiency caused by CD40LG mutations, potentially treatable with CD4+ T-cell gene editing with Cas9 and a "one-size-fits-most" corrective template. Contrary to established gene therapies, there is limited data on the genomic alterations following long-range gene editing, and no consensus on the relevant assays. We developed drop-off digital PCR assays for unbiased detection of large on-target deletions and found them at high frequency upon editing. Large deletions were also common upon editing different loci and cell types and using alternative Cas9 and template delivery methods. In CD40LG edited T cells, on-target deletions were counter-selected in culture and further purged by enrichment for edited cells using a selector coupled to gene correction. We then validated the sensitivity of optical genome mapping for unbiased detection of genome wide rearrangements and uncovered on-target trapping of one or more vector copies, which do not compromise functionality, upon editing using an integrase defective lentiviral donor template. No other recurring events were detected. Edited patient cells showed faithful reconstitution of CD40LG regulated expression and function with a satisfactory safety profile. Large deletions and donor template integrations should be anticipated and accounted for when designing and testing similar gene editing strategies.
Keywords: AAV6; CRISPR; IDLV; large deletions; large integrations.
© 2023 The Authors. Published under the terms of the CC BY NC ND 4.0 license.
Conflict of interest statement
LN, AV, SF, SP, DC, MF and AJ are inventors of patent applications owned by Ospedale San Raffaele and Fondazione Telethon, including one patent application on
Figures

- A
Overview of gene correction strategy. The corrective cassette consists of homology arms (dashed lines), a splice acceptor sequence (SA), the corrective cDNA of exons 2–5, and is followed in its final design by an internal ribosome entry sequence (IRES) cassette that couples reconstitution of CD40LG with surface expression of the low affinity nerve growth factor receptor (LNGFR). It is integrated in intron 1 of the CD40LG gene, targeted by Cas9 ribonucleoprotein.
- B
Experimental design. CD4+ lymphocytes were isolated Day 0 from buffy coats or leukaphereses, cultured for 2–3 days in presence of CD3/CD28 stimulation, IL7 and IL15, and then electroporated and transduced with an adenoviral associated vector serotype 6 (AAV6) or an integrase defective vector (IDLV) on day 2 or 3. Optional enrichment for surface LNGFR took place on day 6; cells were then expanded until day 13–14.
- C
Schematic representation of ddPCR CNV assays' positions on chromosome X with respect to the CD40LG locus (colored boxes).
- D
Loss of LNC00892 amplicons 1–2 days after editing and HDR efficiency 6 days after editing in males (M, n = 11) and female (F, n = 15) from seven independent experiments. Two tailed Mann Whitney test.
- E
Variation over time of LNC00892 copies/diploid genome in males (blue) and females (pink) with respect to untreated samples. Median + IQR. Kruskal–Wallis test for area under the curve, n = 6 males + 6 females.
- F
Expression of LNGFR in patients' cells expressing CD40LG after gene editing and stimulation (n = 4).
- G
Variation in LNC00892 copy number after enrichment (immunomagnetic or sorting), on the day of selection. Pool of five male donors (blue) and six female (pink) donors from 3 independent experiments. Wilcoxon test.
- H–J
Comparison of LNC00892 copies/diploid genome in CD4+ T‐cells edited at the CD40LG locus in absence or presence of AAV6 donor template 1–2 days after editing (n = 4 female and 8 male donors, in pink and blue respectively; (I) RNP cutting efficiency and (J) HDR efficiency, both 6 days after editing). Two tailed Wilcoxon test.
- K, L
Frequency of LNC00892 deletions 1 day after editing with increasing MOI and corresponding HDR efficiency 6 days after editing (K; n = 3 biological replicates).
- M, N
Variation in HDR edited alleles early after editing (M) and corresponding reduction in proportion of deletions in the same samples (N, n = 9 female donors, in pink, and 5 male donors, in blue). Two tailed Wilcoxon test.
- O–Q
Frequency of deletions in IDLV‐edited cells 6 days after editing before and after LNGFR selection (IDLV POS) (n = 6 female donors, in pink and 3 male donors, in blue; two‐tailed Wilcoxon matched‐pairs signed rank test) and corresponding cutting efficiency (P) and HDR efficiency (Q).
- R, S
Copy number variation of target amplicon (AAVS1, B2M, or IL2RG) in colonies derived from cord blood (CB) or mobilized peripheral blood (MPB), placed at 126–800 bp from the DNA DSB. Cas9 was delivered as RNP or RNA. Two sided Fisher's exact test comparing proportion of cells below the cutoff (dashed line).

- A–F
Early timecourse of LNC00892 deletions (n = 2 male and 2 female donors; Median + IQR). Variation over time of ARHGEF6 (B), or MECP2 (C) copies/diploid genome in males (blue) and females (pink) with respect to untreated samples. Median + IQR. Kruskal–Wallis test for area under the curve, n = 5 males + 5 females (B) or n = 6 males + 6 females (C) Editing efficiency by flow cytometry at day 6–9 of Figs 1E, and EV1B and C. Median and range. M, male; F, female; RNP, ribonucleoprotein. (E) Gating strategies for CD40L and NGFR expression (F) Biallelic B2M knock‐out efficiency by flow‐citometry in bulk edited cells.

- A
CD4+ T‐cells from HIGM1 and wild type (WT) mice were primed in vivo against 2,4,6, Trinitrophenyl Keyhole Limpet Hemocyanin (TNP‐KLH). Fourteen days after vaccination CD4+ T‐cells were isolated from the spleen and activated in vitro with anti‐CD3/antiCD28 beads as previously reported (Vavassori et al, 2021). Recipient HIGM1 mice either received 300 mg/kg of cyclophosphamide (CPA) or not, and were transplanted with 107 primed WT CD4+ T‐cells alone or 107 primed WT CD4+ T‐cells admixed with 107 primed HIGM1 CD4+ T‐cells. Mice were vaccinated with TNP‐KLH 21 days and boosted 42 days after CD4+ T cell infusion. Serum was collected 14 days after the first vaccination and 7 days after the second one.
- B–D
Absolute engraftment levels of WT CD4+ T‐cells by flow cytometry (C, D) TNP‐KLH specific (C) and total IgG (D) production after the first vaccination (+14 days) and after boosting (+28 days) in the different subgroups. Gray circles indicate untreated HIGM1 mice, black circles untreated WT mice.

Schematic workflow: (1) A subclone (in red) was generated from bulk SKW 6.4 cell line. (2) A linearized plasmid constitutively expressing GFP was integrated into the IL2RG locus by trapping in a Cas9 induced DSB. A single clone containing the correct insert was characterized and expanded. (3) The same GFP+ clone was cut simultaneously at two 300‐kb spaced loci (GPR112 and ARHFEF6), respectively upstream and downstream the CD40LG locus on chromosome X. A single cell clone was derived again after this modification, characterized, and expanded. (4) The clone of interest (purple) was admixed in known proportions with the unmodified clone (step 1, in red) by single cell sorting. (5) Samples admixed at different proportions (pink‐purple) were compared against their unmodified counterpart derived in step 1 (in red).
Reconstruction of the 6.9 kb insertion in the IL2RG locus. Sequenced junctions are shown by dark red arrows.
Large genomic rearrangements detected in the selected clone. Bars indicate ratios between ddPCR CNV assays located on chromosome X, near the centromere (ARHGEF9), on chromosome X, near the telomere (MECP2), within the rearranged region comprised within the Cas9 target sites (ARHGEF6 and LNC00892), chromosome 14 (TTC5), chromosome 19 (EPS8L1), chromosome 1 (EIF2C1). Bars indicate 95% confidence interval of the copy number.
Comparison of Sample #1, containing 10% of cells with genomic rearrangements of the X chromosome, against Sample #0 consisting of unmodified cells. Circos plot: circles from the most external to the most internal represent: (1) Chromosome number. (2) Conventional banding. (3) Annotated genes. (4) Structural variants, color coded. (5) Copy number variations. (6) Translocations (none present in current plot). The green dot on chromosome X indicates an insertion in the IL2RG locus, while the blue and purple dots indicate a rearrangement around the CD40LG locus.
Integration of the reporter plasmid in the IL2RG locus. Tracks from top to bottom: (1) Banding of the X chromosome. (2) Copy number variations (light green track). (3) Structural variants; the horizontal dark green line indicates an insertion. (4) Annotated genes. (5) Reference assembly (green, labels in blue). (6) Sample assembly (light blue), with labels matching the reference (dark blue) or not (yellow).



- A
LNGFR expression by flow cytometry in CD4+ T‐cells at day 6 (day of immunomagnetic enrichment of LNGFR+ cells) and day 14 (end of process).
- B, C
Results of karyotype analysis performed on three bulk edited biological replicates 14 days after editing and (C) representative images (n = 92, 95 and 91 metaphases respectively for donor 1, 2, and 3).
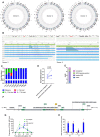
- A
Circos plots of LNGFR+ CD4+ T‐cells compared with each donor cultured unedited counterpart, annotated as in Fig 2D. The green dot on chromosome X indicates an insertion in the CD40LG locus.
- B
Representation of insertion events around CD40LG locus in donor #2 (left) and donor #3 (right). Tracks from top to bottom: (1) Banding of the X chromosome. (2) Copy number variations (green track). (3) Structural variants; green line indicates insertion (also represented by shaded trapezium). (4) Annotated genes. (5) Reference assembly (green, labels in blue). (6) Sample assemblies (light blue, shaded in dark blue according to relative abundance), with labels matching the reference (dark blue) or not (yellow).
- C
Proportion of insertions at CD40LG locus of different color‐coded size in bulk, LNGFR‐depleted (neg) and enriched (pos) fractions of donors 1, 2 and 3.
- D
IDLV VCN in the same LNGFR enriched samples of Figs 3A and 4A and three additional LNGFR enriched samples from similar experiments, before and after digestion with SphI. One‐tailed Wilcoxon matched‐pairs signed rank test.
- E, F
Distribution and absolute number of on target reads from two samples of gene edited CD4+ T‐cells after LNGFR selection and expansion (F) Detailed map of concatemer.
- G, H
Median fluorescent intensuity (MFI) if CD40L in LNGFR+ cells (median‐+IQR). H. CD40 downstream signal transduction by SEAP colorimetric assay (mean ± SEM). Each color represents a different experiment. Pt, Patient; HD, healthy donor; UT, untreated and GE, gene edited.

- A–C
Alignment of amplicons retrieved by nanopore sequencing, in red, to HDR (A) imprecise HDR (B) and native gene (C). gRNA1‐4 indicate the positions of the gRNAs used for target sequence enrichment.
References
-
- Adikusuma F, Piltz S, Corbett MA, Turvey M, McColl SR, Helbig KJ, Beard MR, Hughes J, Pomerantz RT, Thomas PQ (2018) Large deletions induced by Cas9 cleavage. Nature 560: E8–E9 - PubMed
-
- Brown MP, Topham DJ, Sangster MY, Zhao J, Flynn KJ, Surman SL, Woodland DL, Doherty PC, Farr AG, Pattengale PK et al (1998) Thymic lymphoproliferative disease after successful correction of CD40 ligand deficiency by gene transfer in mice. Nat Med 4: 1253–1260 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

