Informed interpretation of metagenomic data by StrainPhlAn enables strain retention analyses of the upper airway microbiome
- PMID: 37916972
- PMCID: PMC10734448
- DOI: 10.1128/msystems.00724-23
Informed interpretation of metagenomic data by StrainPhlAn enables strain retention analyses of the upper airway microbiome
Abstract
The usage of 16S rRNA gene sequencing has become the state-of-the-art method for the characterization of the microbiota in health and respiratory disease. The method is reliable for low biomass samples due to prior amplification of the 16S rRNA gene but has limitations as species and certainly strain identification is not possible. However, the usage of metagenomic tools for the analyses of microbiome data from low biomass samples is not straight forward, and careful optimization is needed. In this work, we show that by validating StrainPhlAn 3 results with the data from bacterial cultures, the strain-level tracking of the respiratory microbiome is feasible despite the high content of host DNA being present when parameters are carefully optimized to fit low biomass microbiomes. This work further proposes that strain retention analyses are feasible, at least for more abundant species. This will help to better understand the longitudinal dynamics of the upper respiratory microbiome during health and disease.
Keywords: bacterial culture; genome analysis; metagenomics; respiratory tract; strain resolution.
Conflict of interest statement
L.S., S.K., and O.S. are employees of Société des Produits Nestlé (SPN). P.L. reports grants (from Vertex and OM Pharma), lecture fees (from Vertex, Vifor, and OM Pharma), and participation on data and safety monitoring boards or advisory boards (for Polyphor, Santhera, Vertex, OM Pharma, Vifor, and Sanofi Avenits), outside the submitted work. M.H. reports a grant (from Pfizer) and participation on advisory boards (for Pfizer and MSD), outside the submitted work.
Figures
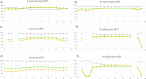


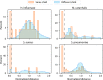

References
-
- Prevaes SMPJ, de Winter-de Groot KM, Janssens HM, de Steenhuijsen Piters WAA, Tramper-Stranders GA, Wyllie AL, Hasrat R, Tiddens HA, van Westreenen M, van der Ent CK, Sanders EAM, Bogaert D. 2016. Development of the nasopharyngeal microbiota in infants with cystic fibrosis. Am J Respir Crit Care Med 193:504–515. doi: 10.1164/rccm.201509-1759OC - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
