Targeting the non-coding genome and temozolomide signature enables CRISPR-mediated glioma oncolysis
- PMID: 37917583
- PMCID: PMC10725516
- DOI: 10.1016/j.celrep.2023.113339
Targeting the non-coding genome and temozolomide signature enables CRISPR-mediated glioma oncolysis
Abstract
Glioblastoma (GBM) is the most common lethal primary brain cancer in adults. Despite treatment regimens including surgical resection, radiotherapy, and temozolomide (TMZ) chemotherapy, growth of residual tumor leads to therapy resistance and death. At recurrence, a quarter to a third of all gliomas have hypermutated genomes, with mutational burdens orders of magnitude greater than in normal tissue. Here, we quantified the mutational landscape progression in a patient's primary and recurrent GBM, and we uncovered Cas9-targetable repeat elements. We show that CRISPR-mediated targeting of highly repetitive loci enables rapid elimination of GBM cells, an approach we term "genome shredding." Importantly, in the patient's recurrent GBM, we identified unique repeat sequences with TMZ mutational signature and demonstrated that their CRISPR targeting enables cancer-specific cell ablation. "Cancer shredding" leverages the non-coding genome and therapy-induced mutational signatures for targeted GBM cell depletion and provides an innovative paradigm to develop treatments for hypermutated glioma.
Keywords: CP: Cancer; CRISPR-Cas9; cancer shredding; genome shredding; glioblastoma; hypermutated glioma.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The Regents of the University of California and the J. David Gladstone Institutes have filed a patent related to this work, on which C.F. is listed as inventor. Silico Therapeutics has filed a patent related to this work, on which A.R.P. and X.S. are listed as inventors. J.A.D. is a co-founder of Caribou Biosciences, Editas Medicine, Intellia Therapeutics, Scribe Therapeutics, and Mammoth Biosciences. J.A.D. is a scientific advisory board member of Caribous Biosciences, Intellia Therapeutics, eFFECTOR Therapeutics, Scribe Therapeutics, Synthego, Metagenomi, Mammoth Biosciences, and Inari. J.A.D. is a member of the board of directors at Driver and Johnson & Johnson. C.F. is a co-founder of Mirimus, Inc.
Figures



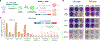
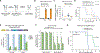


References
-
- Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, and Ellison DW (2016). The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 131, 803–820. 10.1007/s00401-016-1545-1. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

